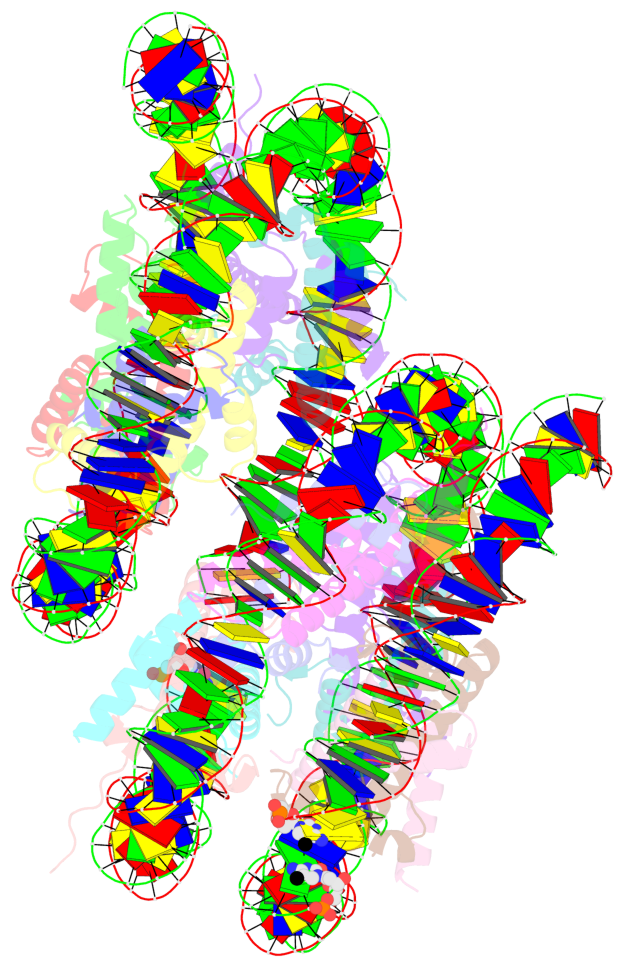
Summary information and primary citation
- PDB-id
-
5gse;
DSSR-derived features in text and
JSON formats; DNAproDB
- Class
- structural protein-DNA
- Method
- X-ray (3.14 Å)
- Summary
- Crystal structure of unusual nucleosome
- Reference
-
Kato D, Osakabe A, Arimura Y, Mizukami Y, Horikoshi N,
Saikusa K, Akashi S, Nishimura Y, Park SY, Nogami J,
Maehara K, Ohkawa Y, Matsumoto A, Kono H, Inoue R,
Sugiyama M, Kurumizaka H (2017): "Crystal
structure of the overlapping dinucleosome composed of
hexasome and octasome." Science,
356, 205-208. doi: 10.1126/science.aak9867.
- Abstract
- Nucleosomes are dynamic entities that are repositioned
along DNA by chromatin remodeling processes. A nucleosome
repositioned by the switch-sucrose nonfermentable (SWI/SNF)
remodeler collides with a neighbor and forms the
intermediate "overlapping dinucleosome." Here, we report
the crystal structure of the overlapping dinucleosome, in
which two nucleosomes are associated, at 3.14-angstrom
resolution. In the overlapping dinucleosome structure, the
unusual "hexasome" nucleosome, composed of the histone
hexamer lacking one H2A-H2B dimer from the conventional
histone octamer, contacts the canonical "octasome"
nucleosome, and they intimately associate. Consequently,
about 250 base pairs of DNA are left-handedly wrapped in
three turns, without a linker DNA segment between the
hexasome and octasome moieties. The overlapping
dinucleosome structure may provide important information to
understand how nucleosome repositioning occurs during the
chromatin remodeling process.
List of 1 5mC-amino acid contact
- The contacts include paired nucleotides (mostly a G in
Watson-Crick G-C pairing) and amino-acids within a 4.5-Å
distance cutoff to base atoms of 5mC.
- The structure is oriented in the base reference frame
of 5mC, allowing for easy comparison and direct
superimposition between entries.
- The black sphere (•) denotes the
5-methyl carbon atom in 5mC.
No. 1 I.5CM27: other-contacts
is-WC-paired is-in-duplex [+]:CcT/AGG