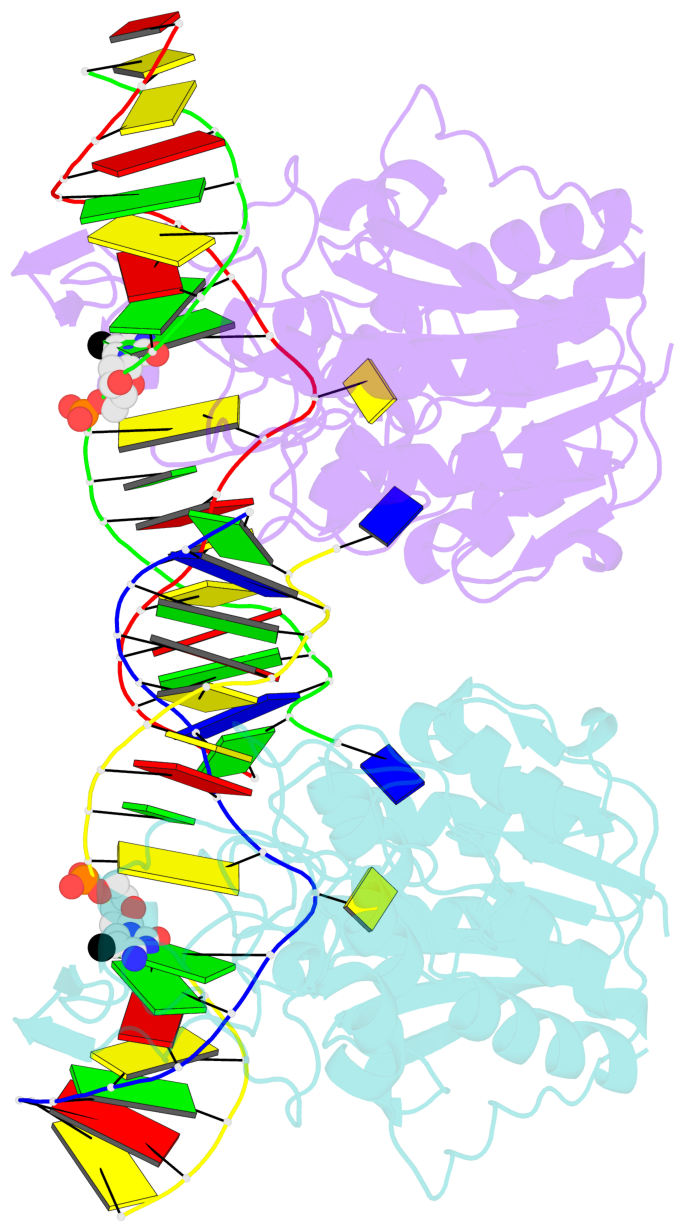
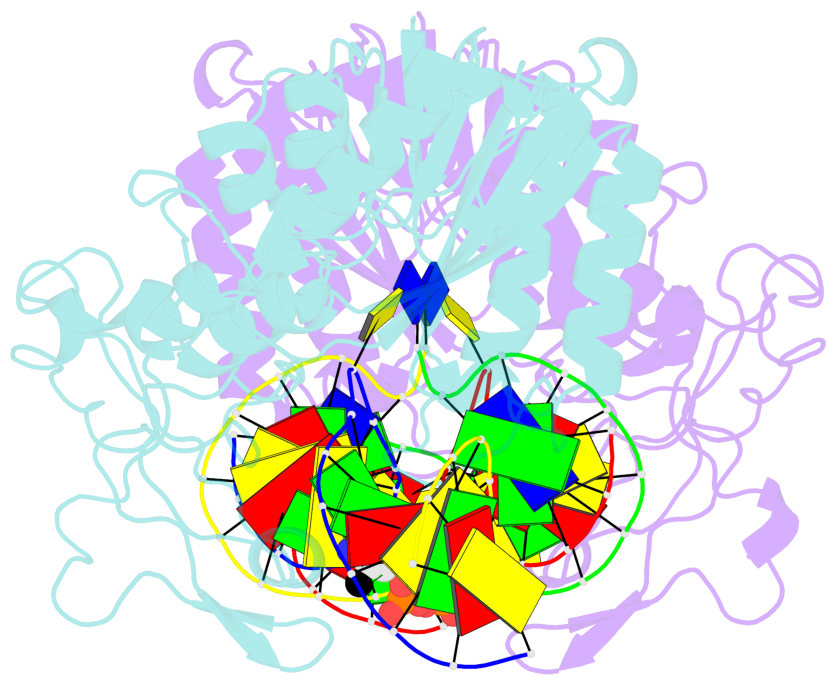
Summary information and primary citation
- PDB-id
- 1dct; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.8 Å)
- Summary
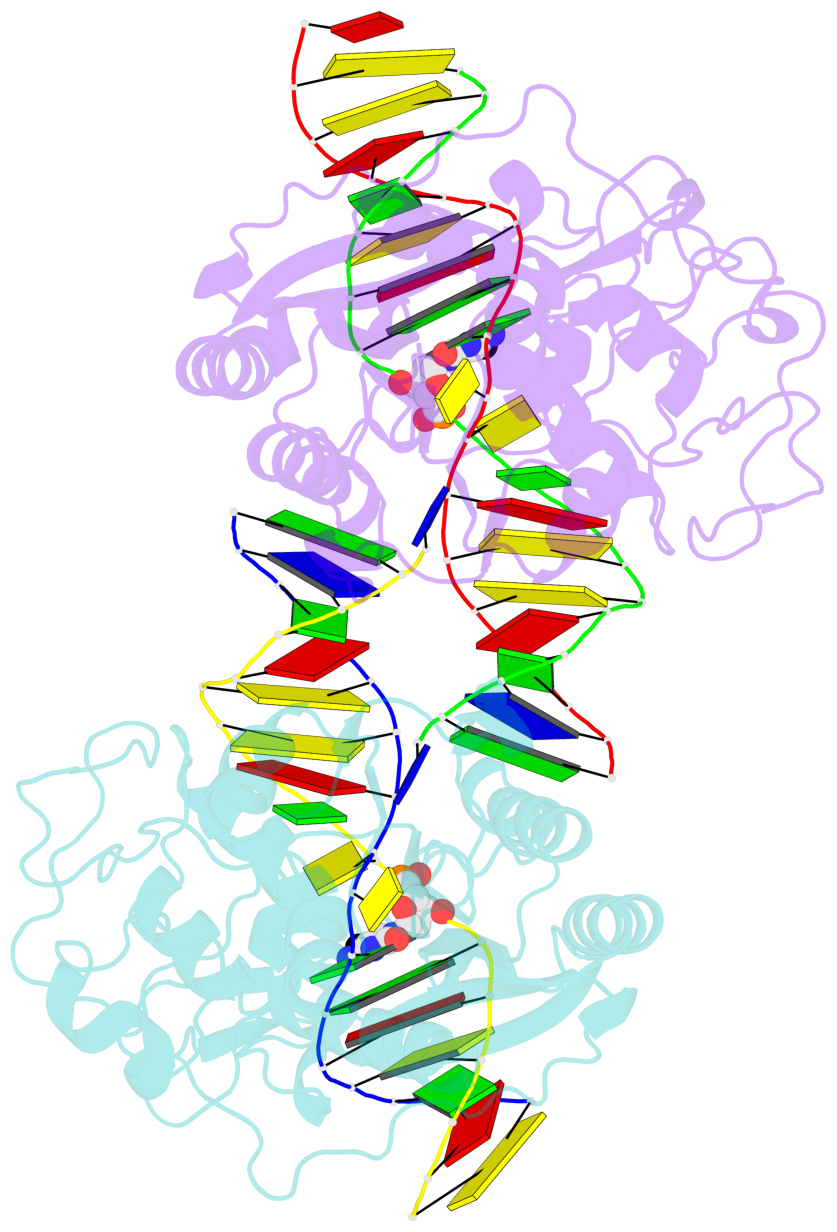
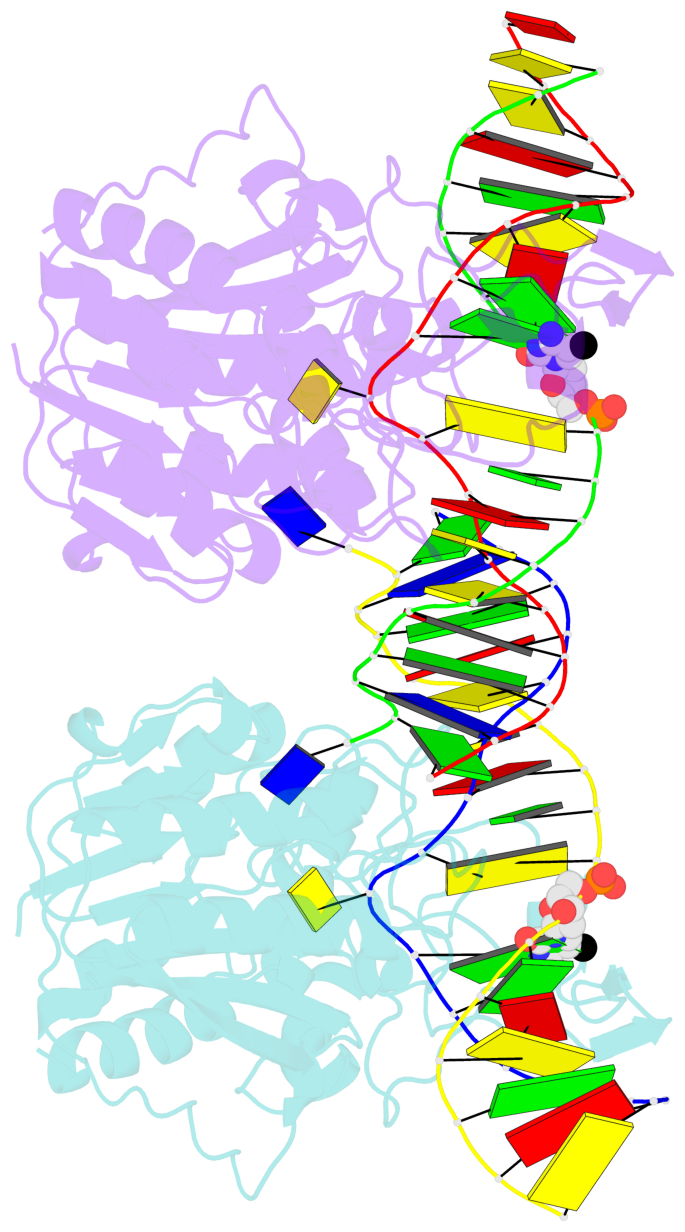
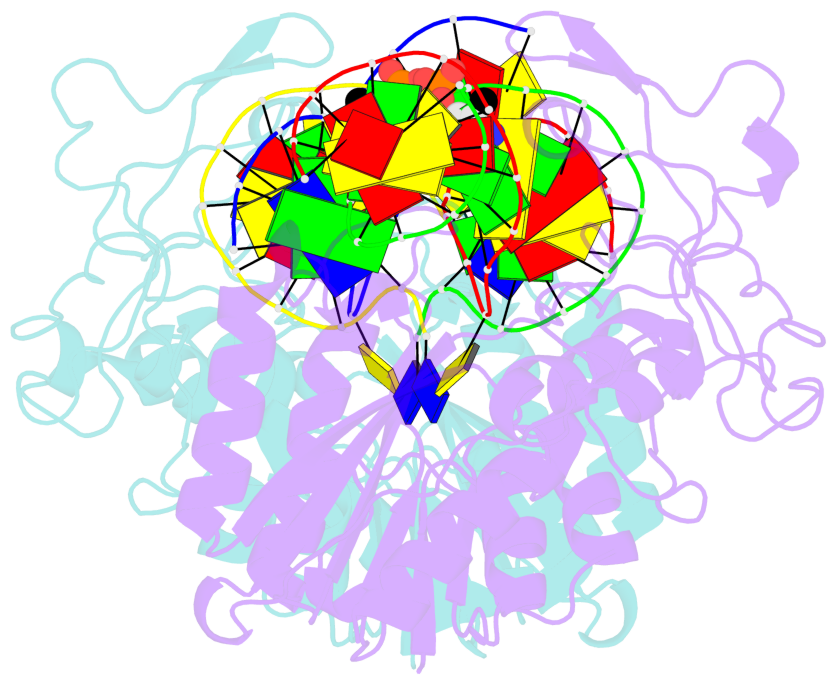
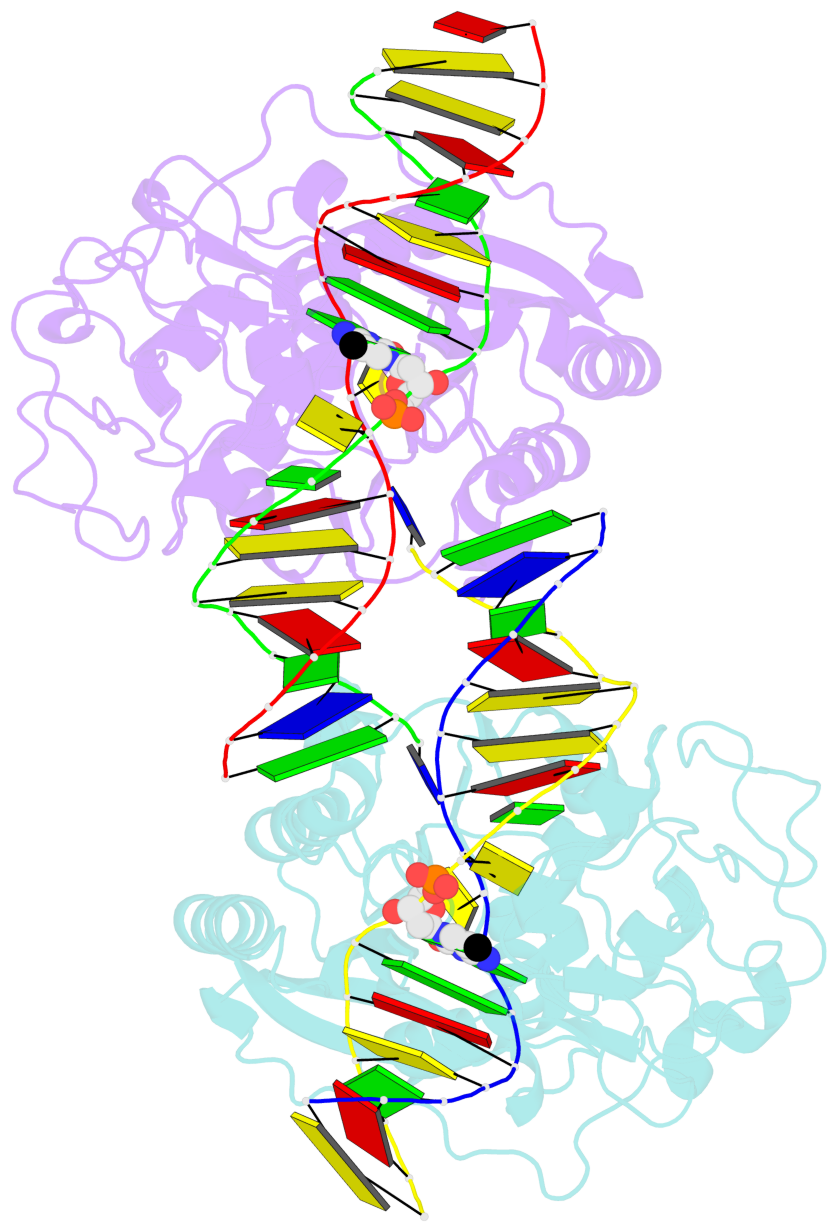
- DNA (cytosine-5) methylase from haeiii covalently bound to DNA
- Reference
- Reinisch KM, Chen L, Verdine GL, Lipscomb WN (1995): "The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing." Cell(Cambridge,Mass.), 82, 143-153. doi: 10.1016/0092-8674(95)90060-8.
- Abstract
- Many organisms expand the information content of their genome through enzymatic methylation of cytosine residues. Here we report the 2.8 A crystal structure of a bacterial DNA (cytosine-5)-methyltransferase (DCMtase), M. HaeIII, bound covalently to DNA. In this complex, the substrate cytosine is extruded from the DNA helix and inserted into the active site of the enzyme, as has been observed for another DCMtase, M. HhaI. The DNA is bound in a cleft between the two domains of the protein and is distorted from the characteristic B-form conformation at its recognition sequence. A comparison of structures shows a variation in the mode of DNA recognition: M. HaeIII differs from M. HhaI in that the remaining bases in its recognition sequence undergo an extensive rearrangement in their pairing. In this process, the bases are unstacked, and a gap 8 A long opens in the DNA.
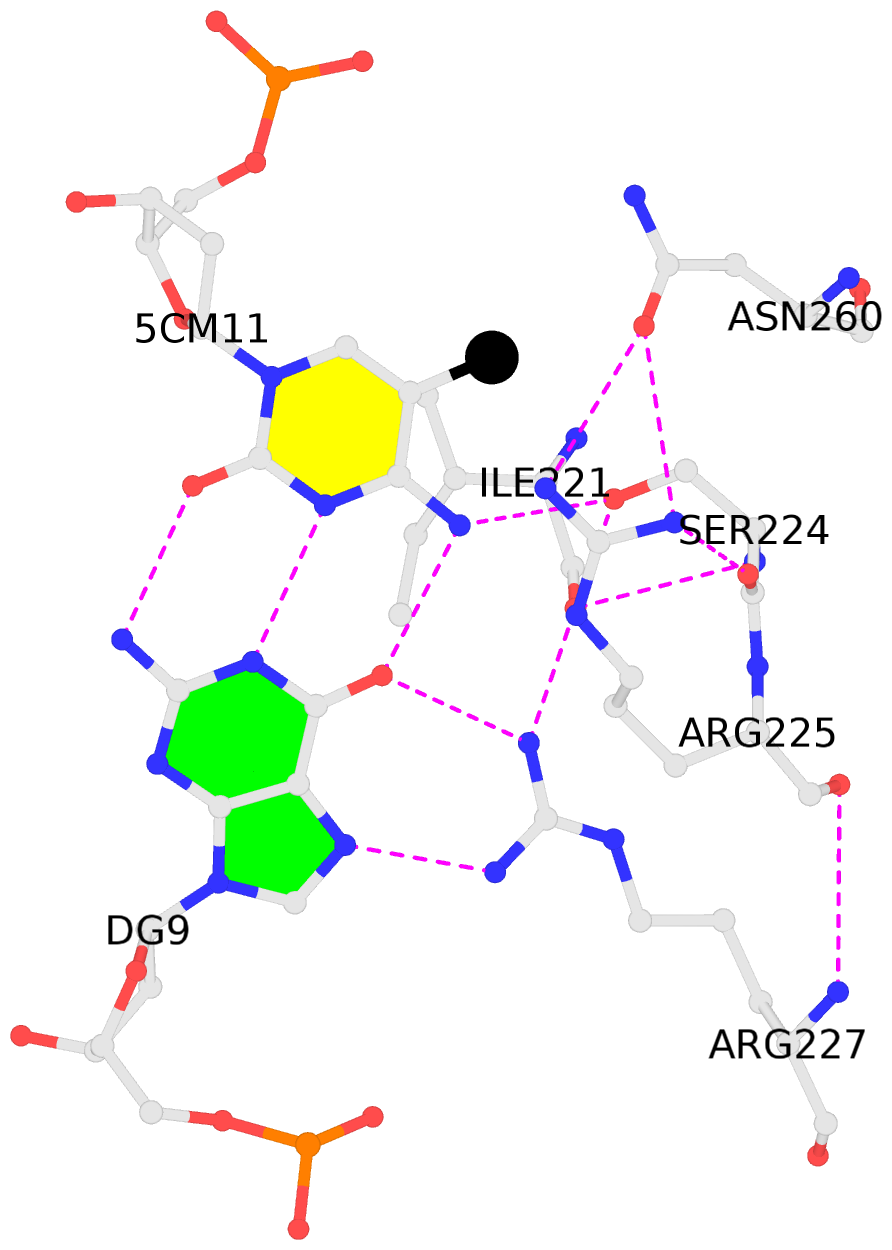
- The contacts include paired nucleotides (mostly a G in Watson-Crick G-C pairing), and
amino-acids within a 4.5-A distance cutoff to base atoms of 5mC.
- The structure is oriented in the base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
No. 1 M.5CM11: hydrophobic-with-A.ILE221 is-WC-paired is-in-duplex [-]:GGC/GcC |
 |
|
No. 2 N.5CM11: stacking-with-B.ARG225 is-WC-paired is-in-duplex [-]:GGC/GcC |
 |
|