Summary information and primary citation
- PDB-id
- 3c2i; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription regulator
- Method
- X-ray (2.5 Å)
- Summary
- The crystal structure of methyl-cpg binding domain of human mecp2 in complex with a methylated DNA sequence from bdnf
- Reference
- Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD (2008): "MeCP2 binding to DNA depends upon hydration at methyl-CpG." Mol.Cell, 29, 525-531. doi: 10.1016/j.molcel.2007.12.028.
- Abstract
- MeCP2 is an essential transcriptional repressor that mediates gene silencing through binding to methylated DNA. Binding specificity has been thought to depend on hydrophobic interactions between cytosine methyl groups and a hydrophobic patch within the methyl-CpG-binding domain (MBD). X-ray analysis of a methylated DNA-MBD cocrystal reveals, however, that the methyl groups make contact with a predominantly hydrophilic surface that includes tightly bound water molecules. This suggests that MeCP2 recognizes hydration of the major groove of methylated DNA rather than cytosine methylation per se. The MeCP2-DNA complex also identifies a unique structural role for T158, the residue most commonly mutated in Rett syndrome.
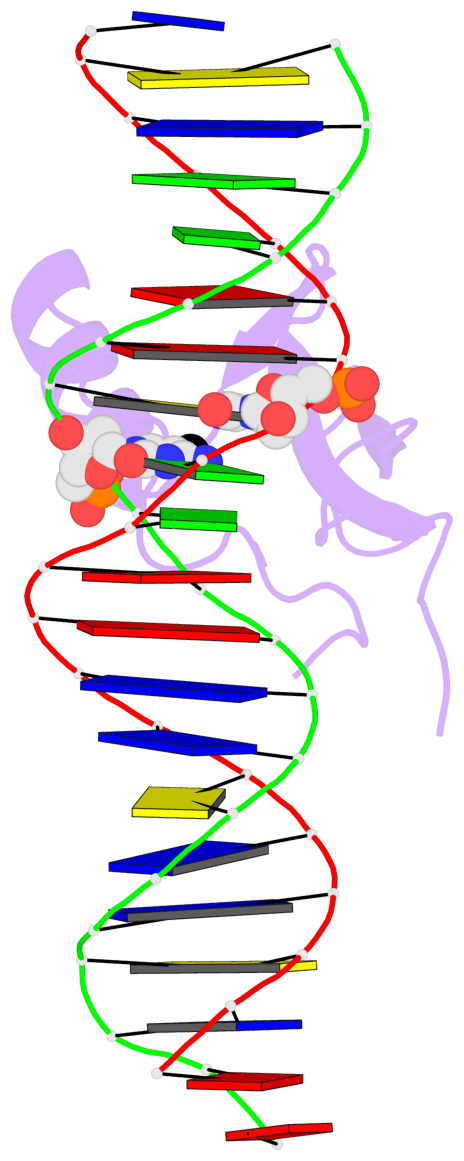
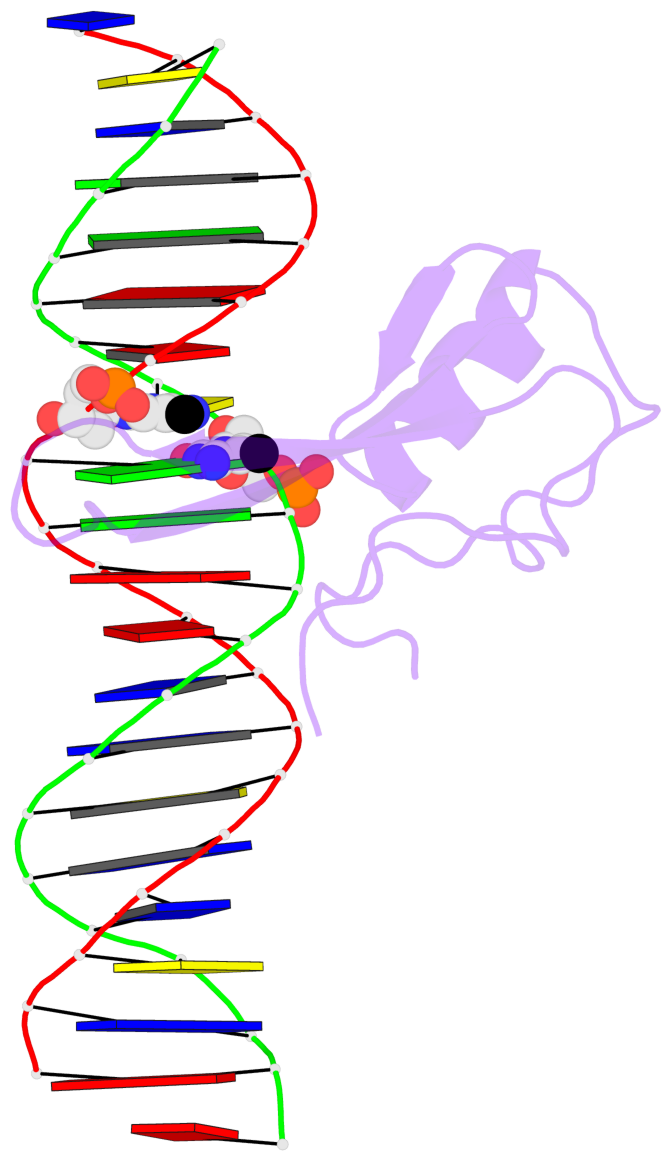
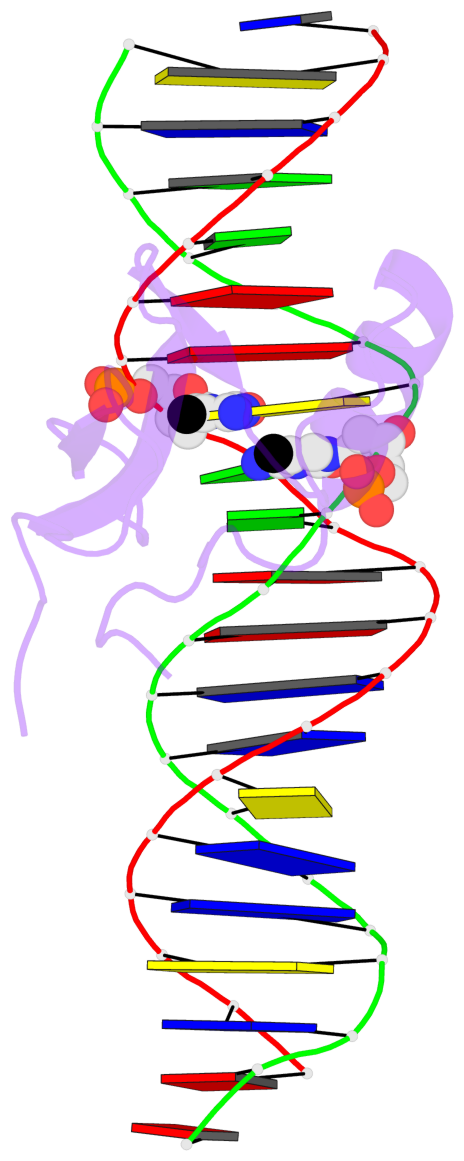
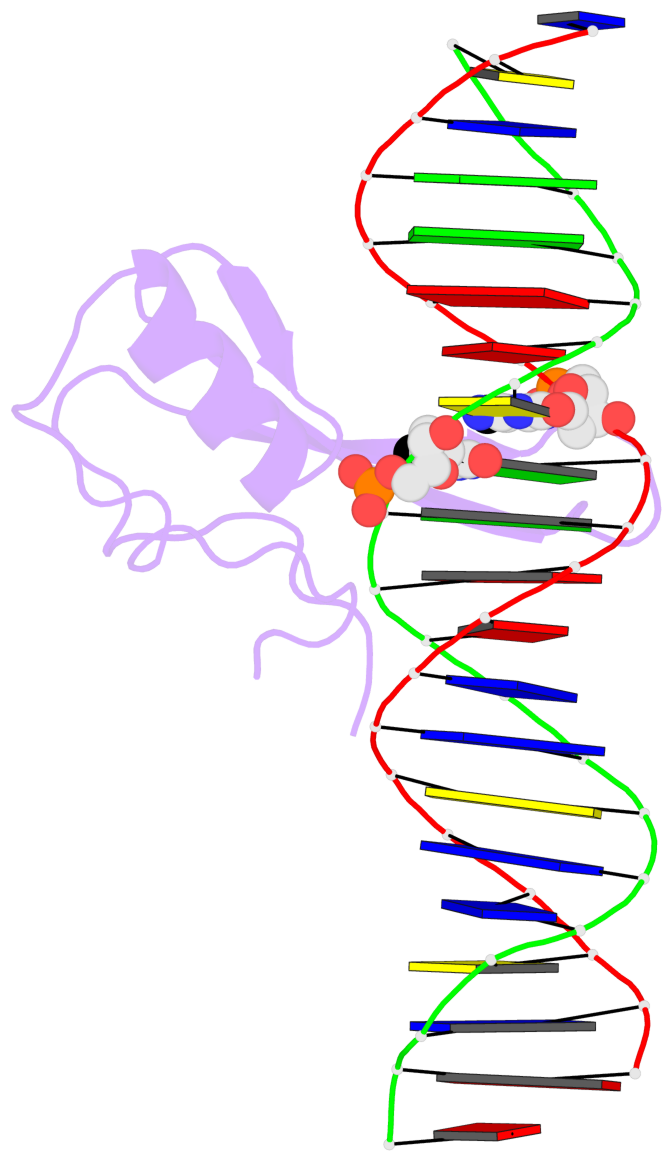
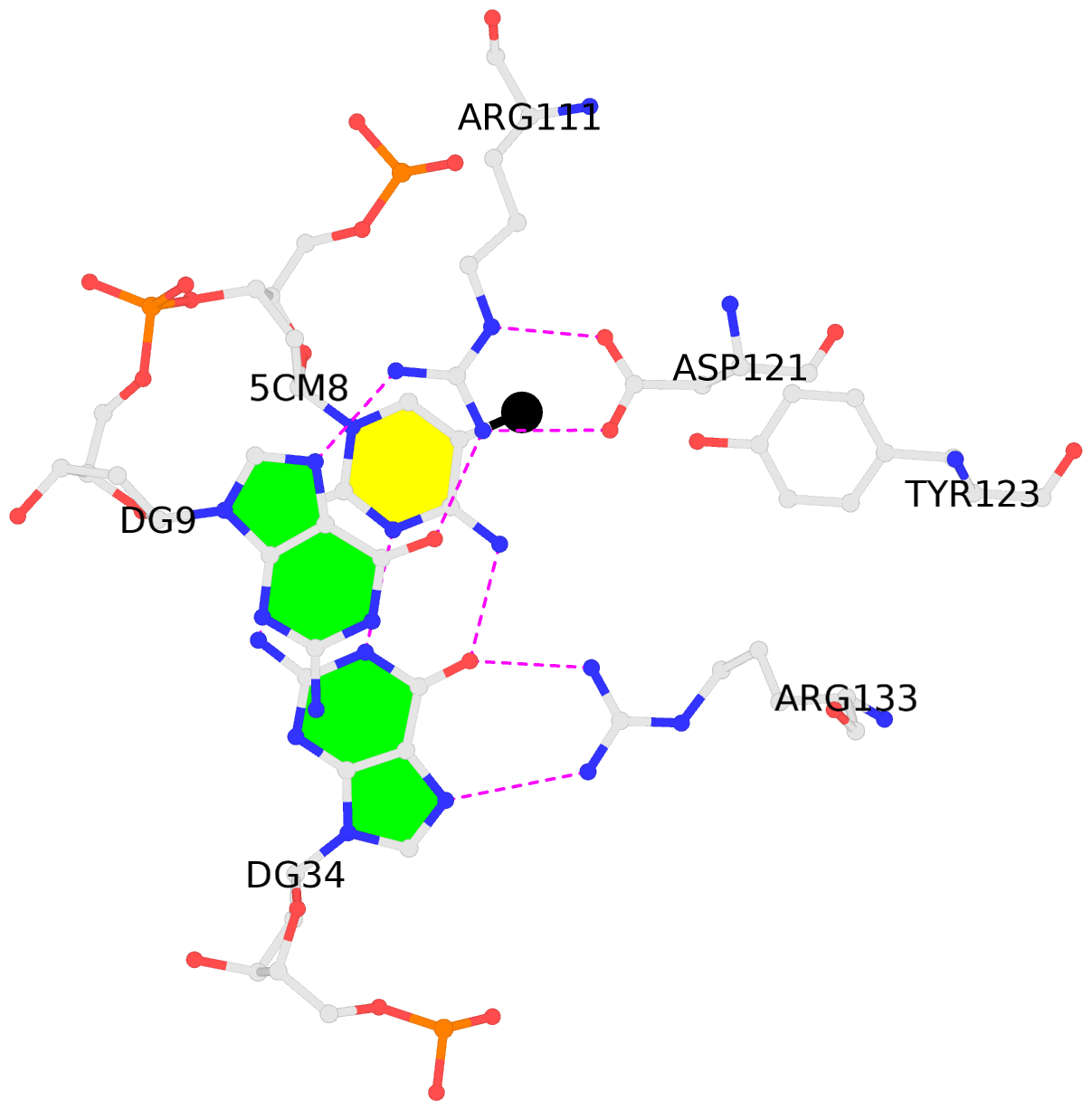
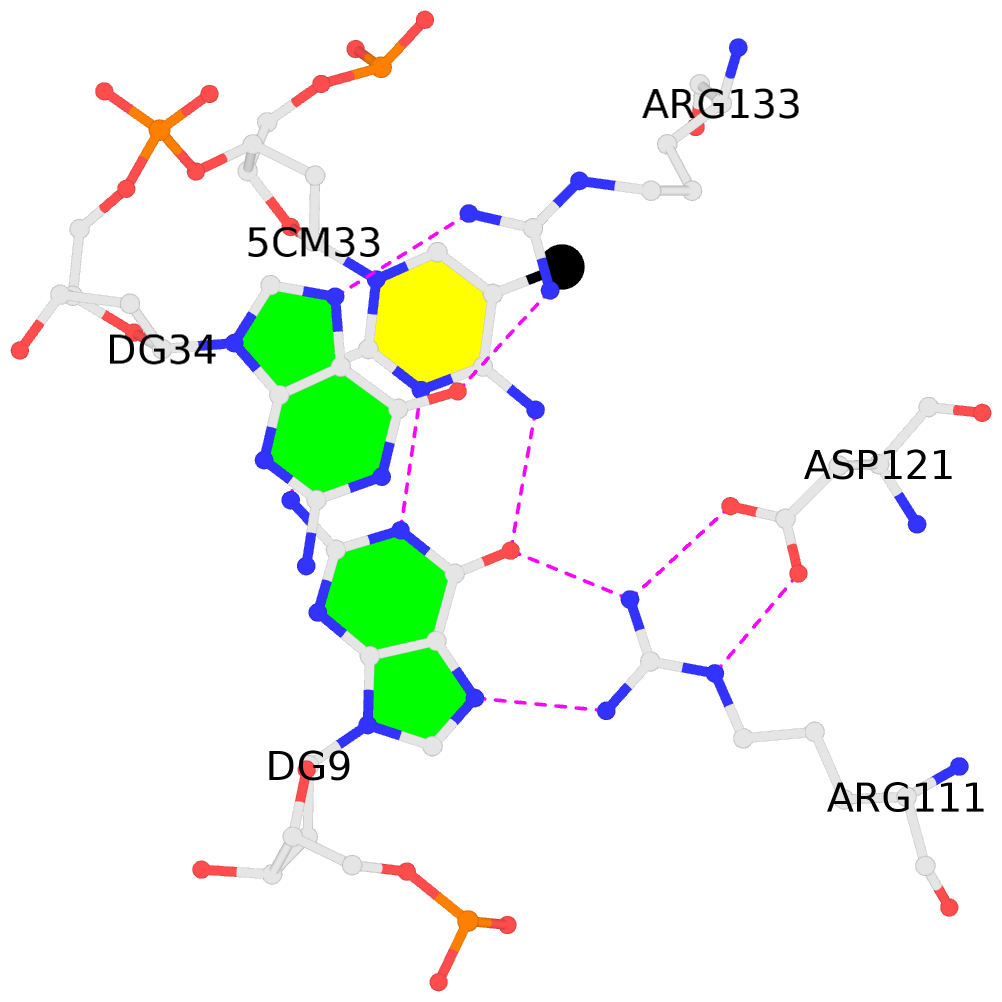
- The contacts include paired nucleotides (mostly a G in Watson-Crick G-C pairing), and
amino-acids within a 4.5-A distance cutoff to base atoms of 5mC.
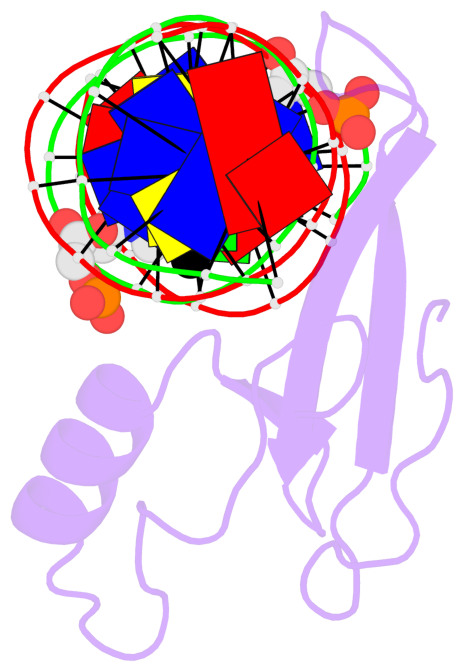
- The structure is oriented in the base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
No. 1 B.5CM8: stacking-with-A.ARG111 is-WC-paired is-in-duplex [+]:AcG/cGT |
 |
|
No. 2 C.5CM33: stacking-with-A.ARG133 is-WC-paired is-in-duplex [-]:cGG/CcG |
 |
|