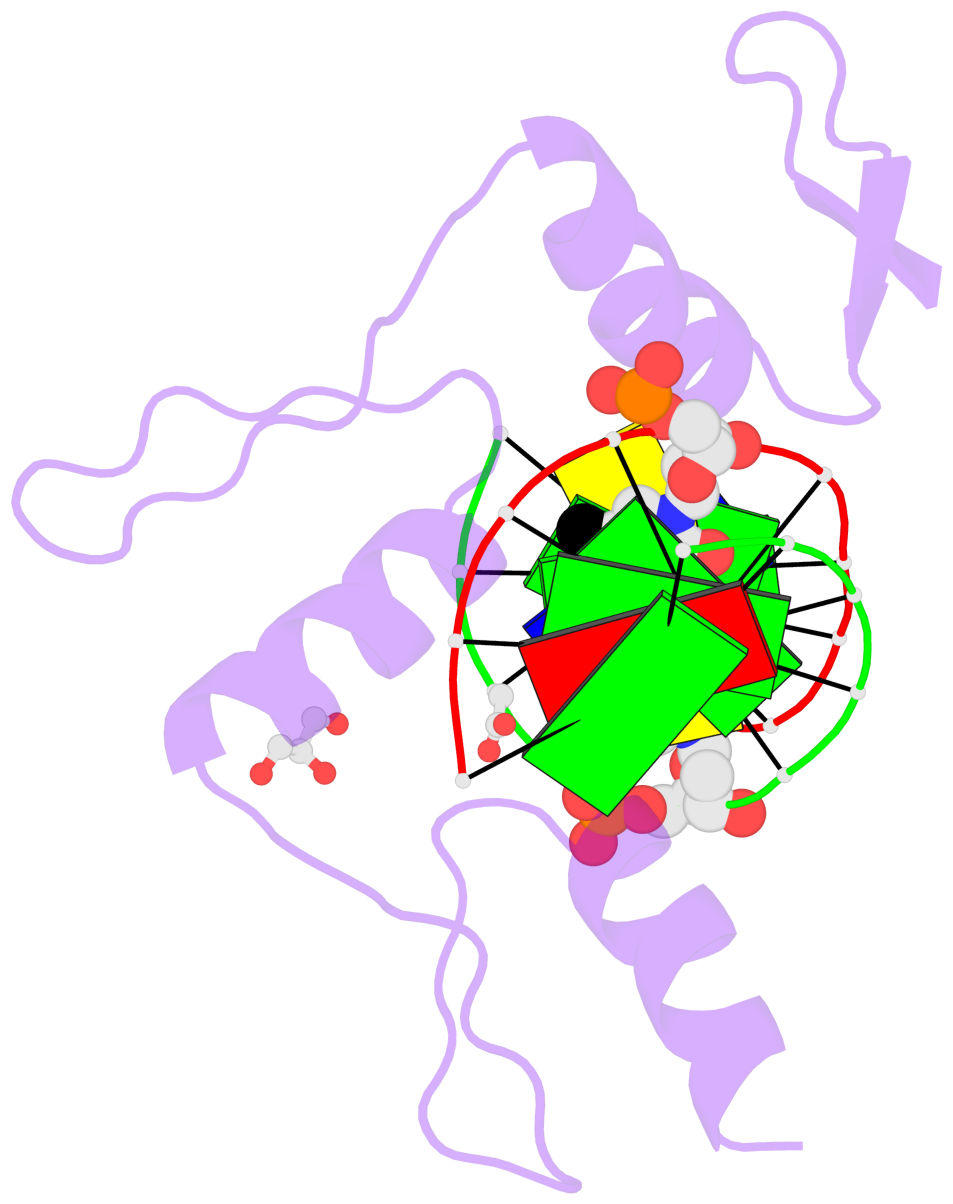
Summary information and primary citation
- PDB-id
- 4m9e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (1.851 Å)
- Summary
- Structure of klf4 zinc finger DNA binding domain in complex with methylated DNA
- Reference
- Liu Y, Olanrewaju YO, Zheng Y, Hashimoto H, Blumenthal RM, Zhang X, Cheng X (2014): "Structural basis for Klf4 recognition of methylated DNA." Nucleic Acids Res., 42, 4859-4867. doi: 10.1093/nar/gku134.
- Abstract
- Transcription factor Krüppel-like factor 4 (Klf4), one of the factors directing cellular reprogramming, recognizes the CpG dinucleotide (whether methylated or unmodified) within a specific G/C-rich sequence. The binding affinity of the mouse Klf4 DNA-binding domain for methylated DNA is only slightly stronger than that for an unmodified oligonucleotide. The structure of the C-terminal three Krüppel-like zinc fingers (ZnFs) of mouse Klf4, in complex with fully methylated DNA, was determined at 1.85 Å resolution. An arginine and a glutamate interact with the methyl group. By comparison with two other recently characterized structures of ZnF protein complexes with methylated DNA, we propose a common principle of recognition of methylated CpG by C2H2 ZnF proteins, which involves a spatially conserved Arg-Glu pair.
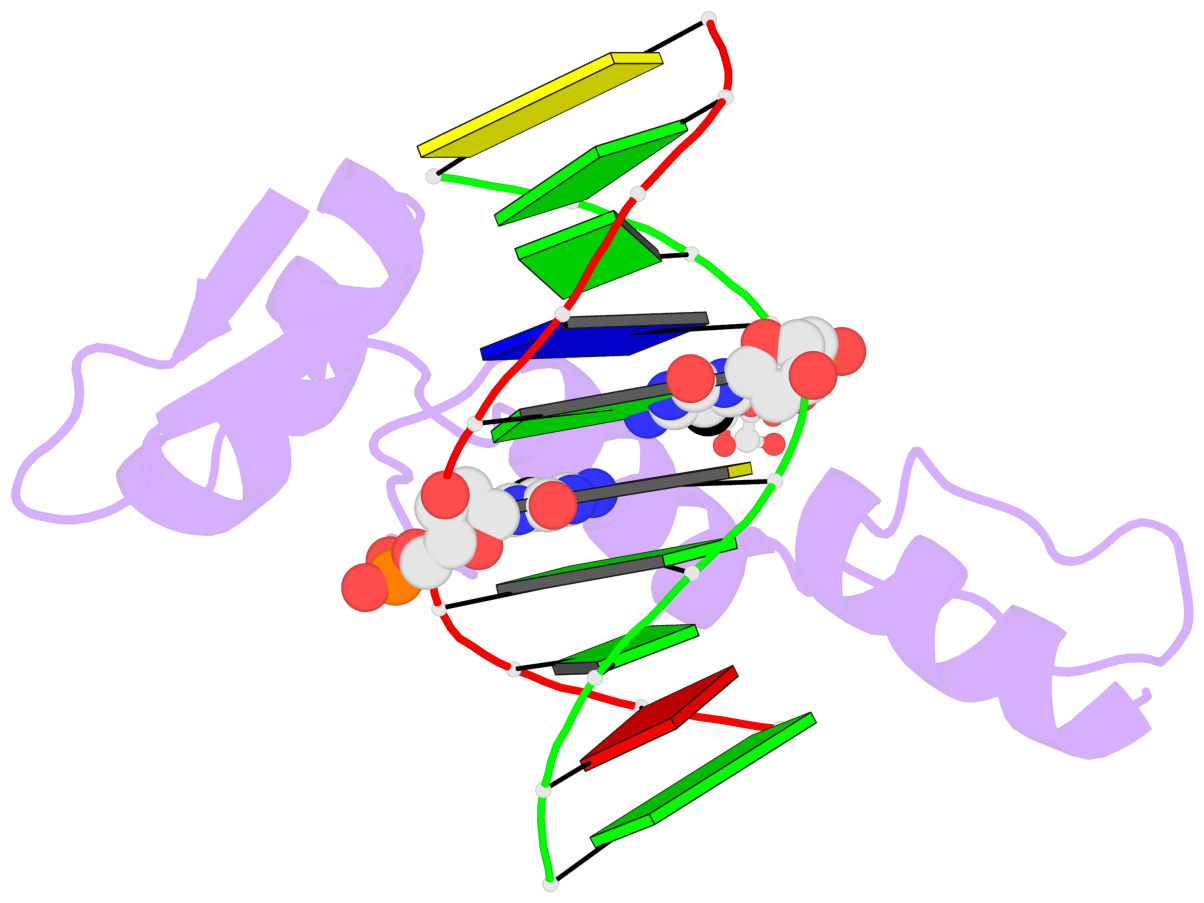
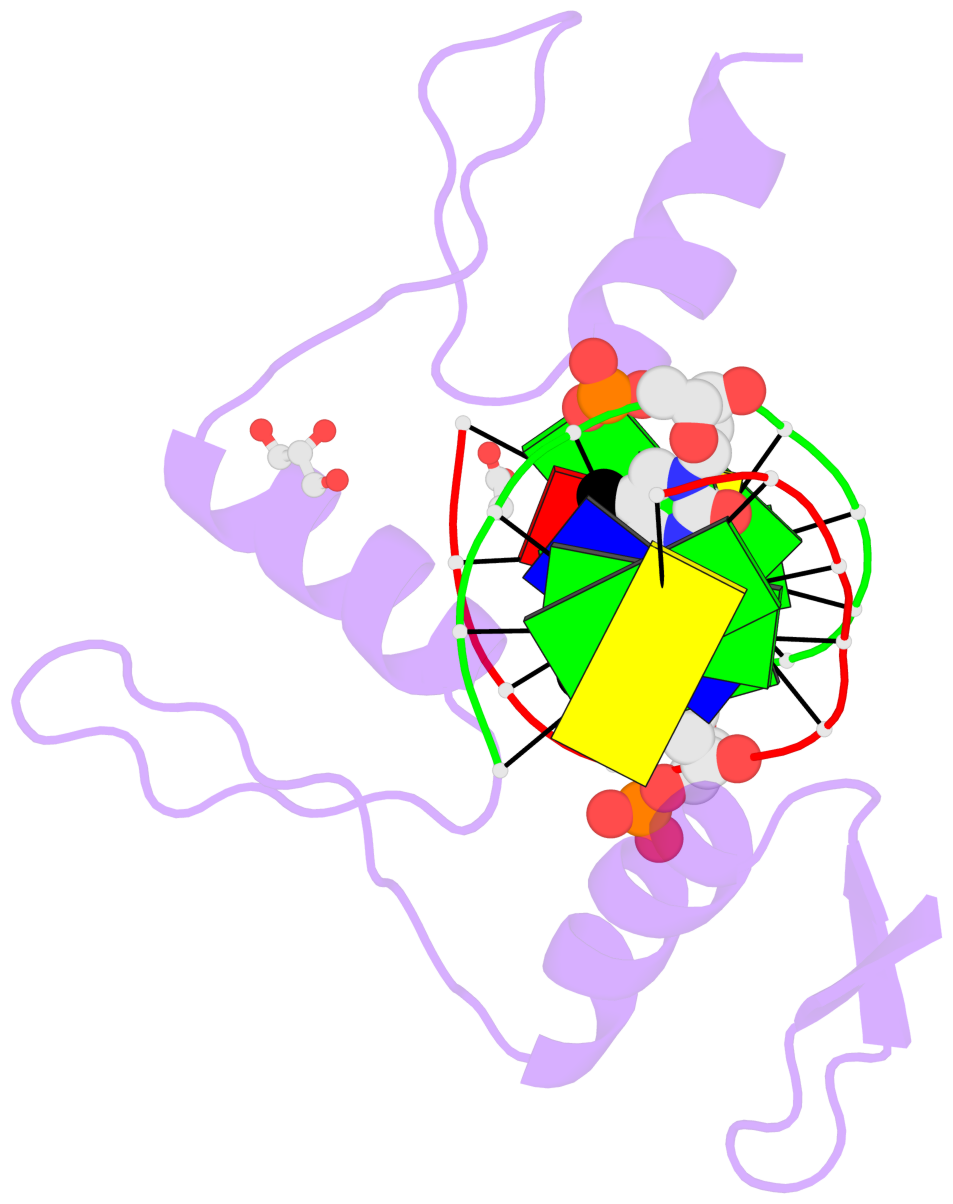
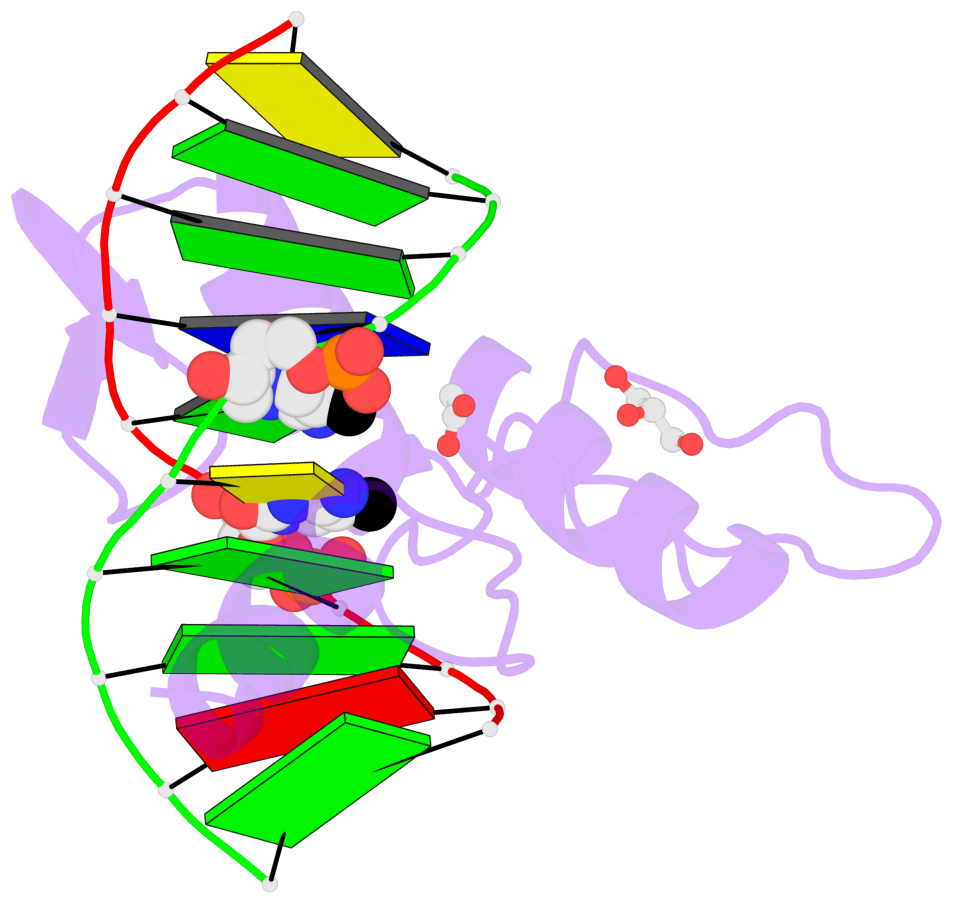
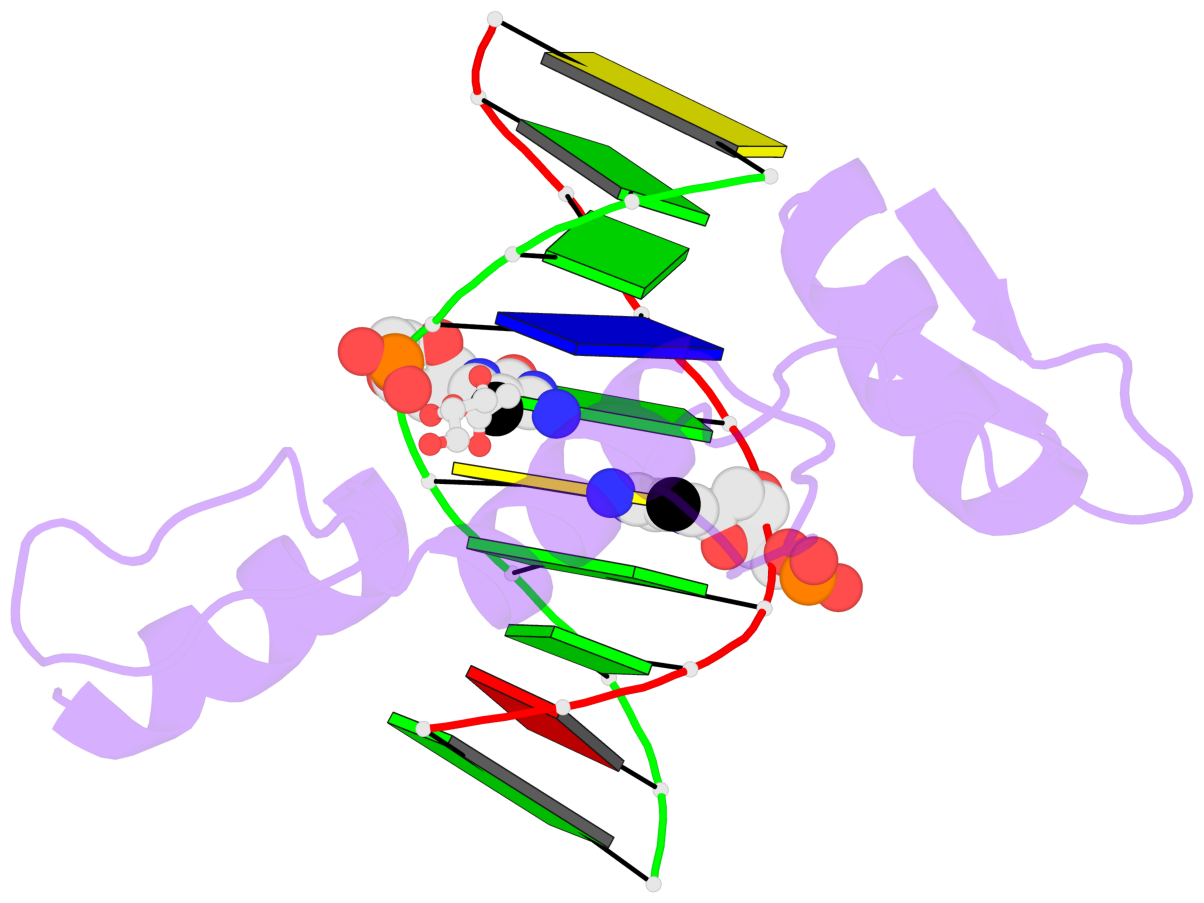
- The contacts include paired nucleotides (mostly a G in Watson-Crick G-C pairing), and
amino-acids within a 4.5-A distance cutoff to base atoms of 5mC.
- The structure is oriented in the base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
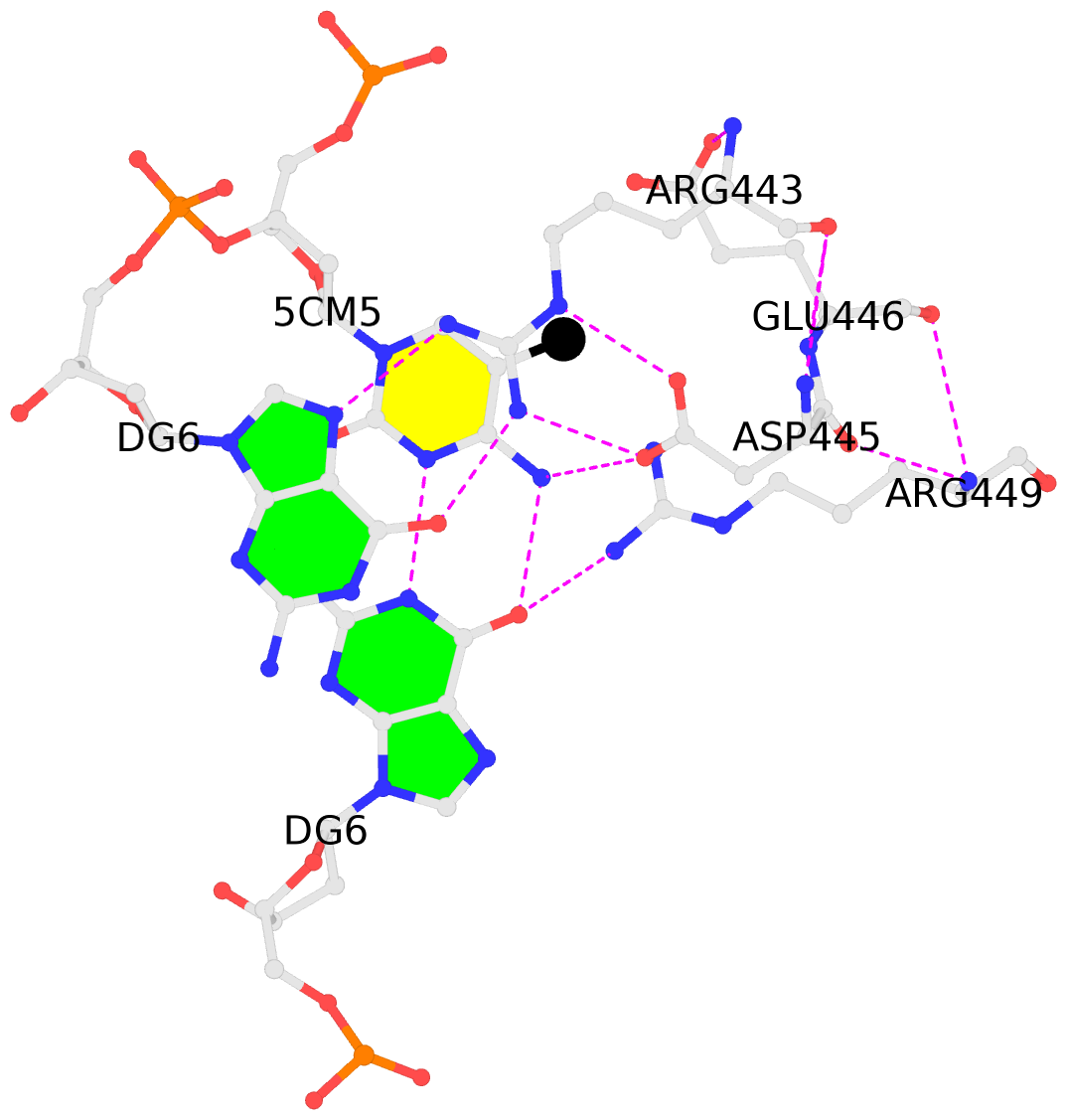
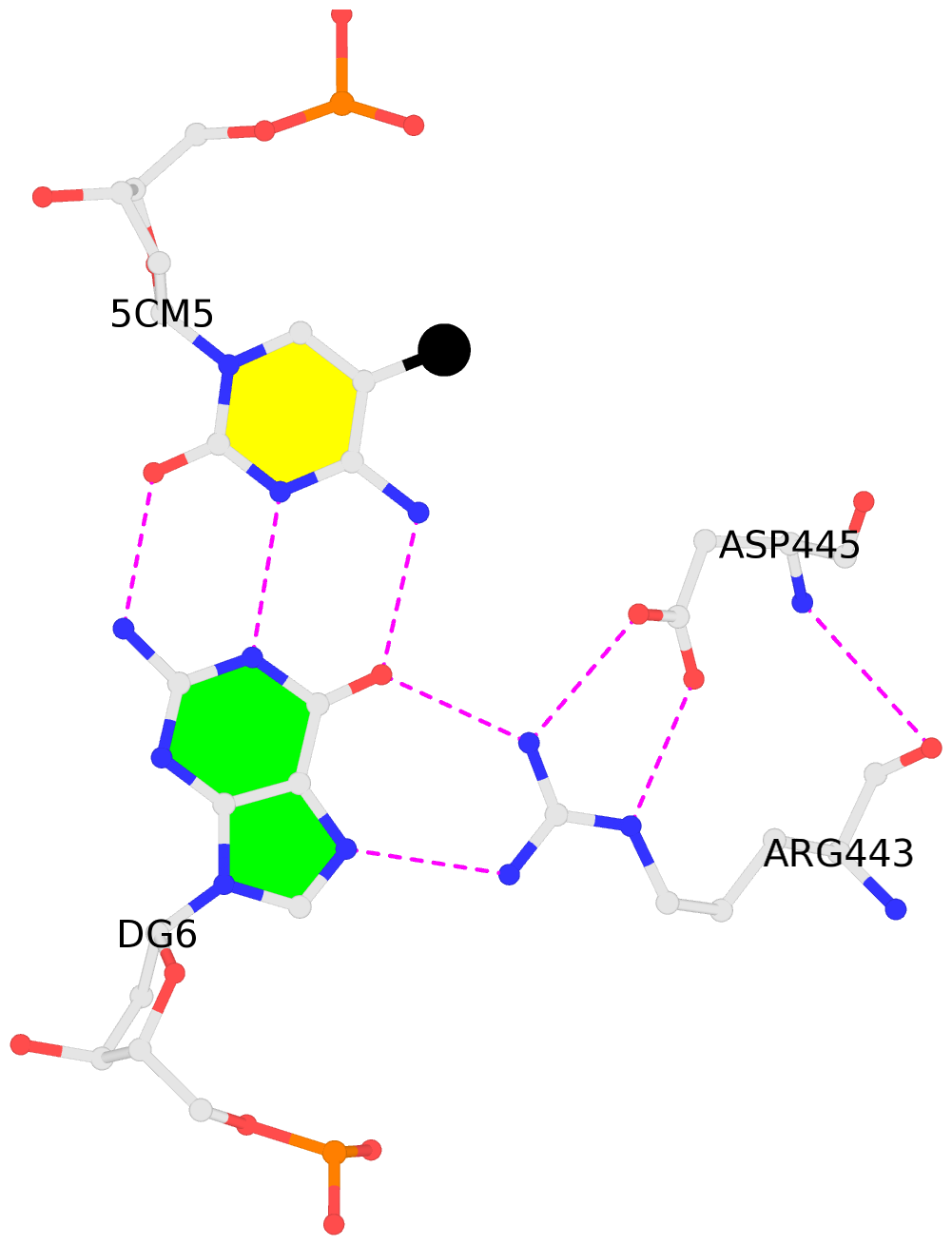
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
No. 1 B.5CM5: stacking-with-A.ARG443 is-WC-paired is-in-duplex [+]:GcG/cGC |
 |
|
No. 2 C.5CM5: other-contacts is-WC-paired is-in-duplex [-]:cGT/AcG |
 |
|