Summary information and primary citation
- PDB-id
- 4nm6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- oxidoreductase-DNA
- Method
- X-ray (2.026 Å)
- Summary
- Crystal structure of tet2-DNA complex
- Reference
- Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y (2013): "Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation." Cell(Cambridge,Mass.), 155, 1545-1555. doi: 10.1016/j.cell.2013.11.020.
- Abstract
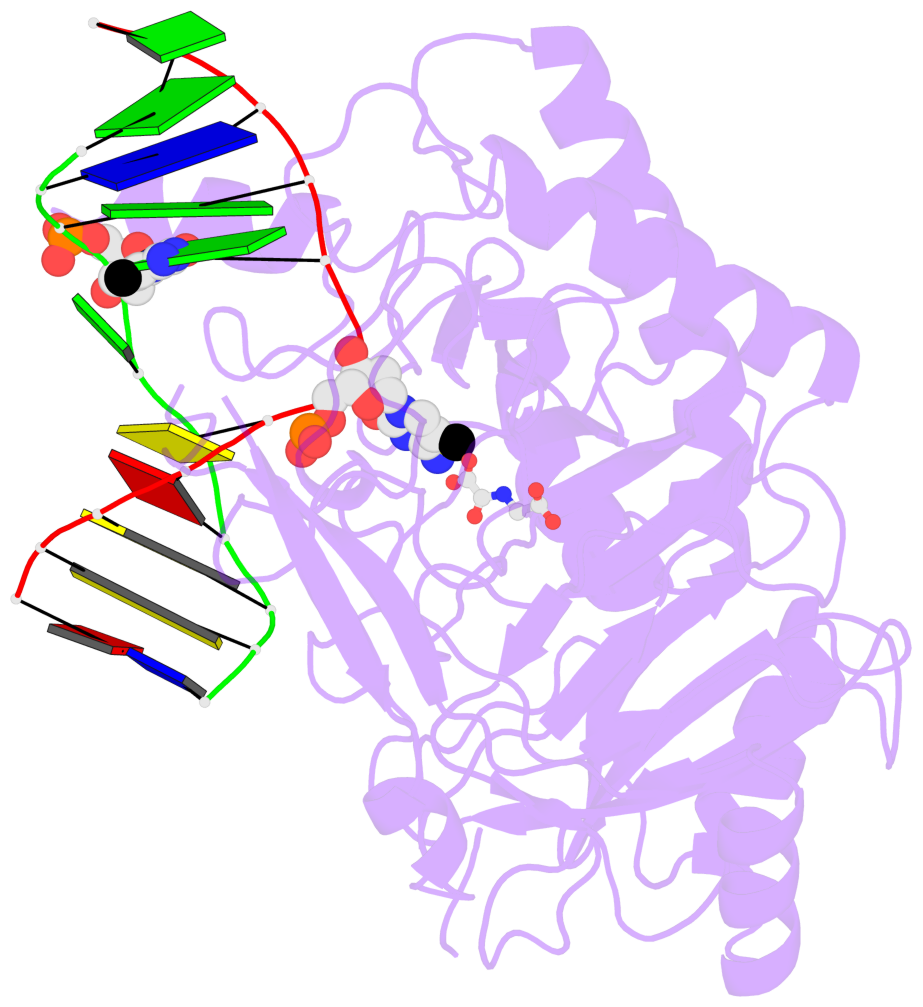
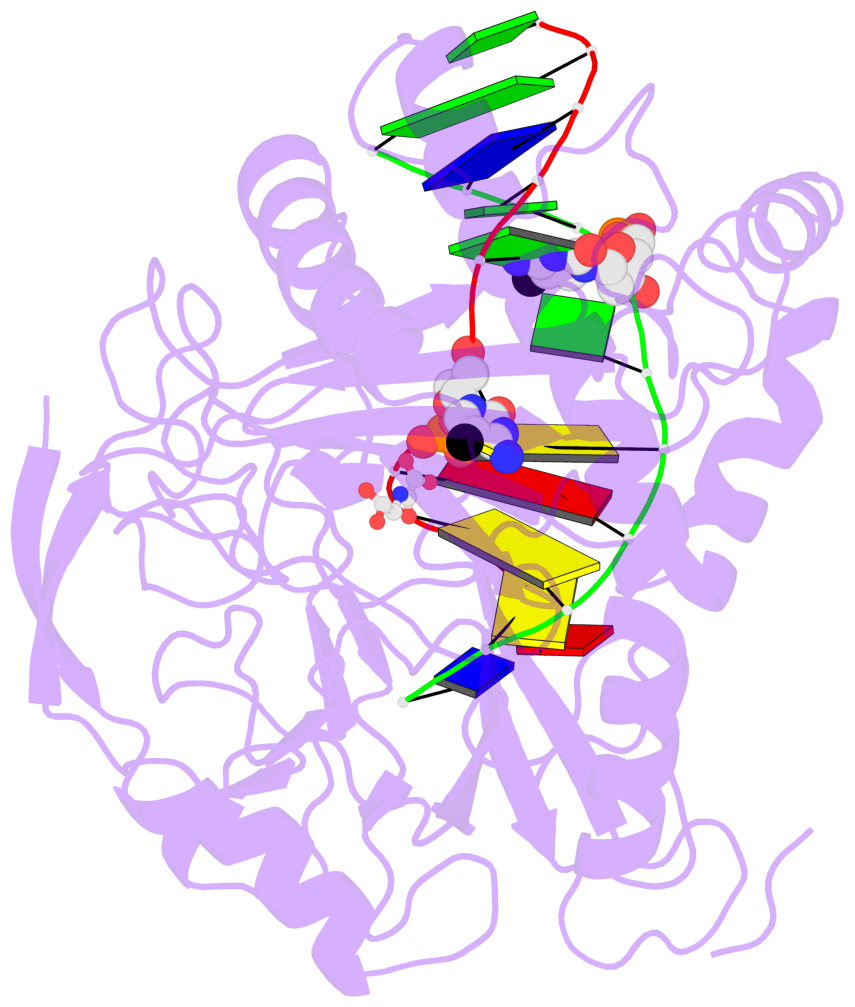
- TET proteins oxidize 5-methylcytosine (5mC) on DNA and play important roles in various biological processes. Mutations of TET2 are frequently observed in myeloid malignance. Here, we present the crystal structure of human TET2 bound to methylated DNA at 2.02 Å resolution. The structure shows that two zinc fingers bring the Cys-rich and DSBH domains together to form a compact catalytic domain. The Cys-rich domain stabilizes the DNA above the DSBH core. TET2 specifically recognizes CpG dinucleotide and shows substrate preference for 5mC in a CpG context. 5mC is inserted into the catalytic cavity with the methyl group orientated to catalytic Fe(II) for reaction. The methyl group is not involved in TET2-DNA contacts so that the catalytic cavity allows TET2 to accommodate 5mC derivatives for further oxidation. Mutations of Fe(II)/NOG-chelating, DNA-interacting, and zinc-chelating residues are frequently observed in human cancers. Our studies provide a structural basis for understanding the mechanisms of TET-mediated 5mC oxidation.
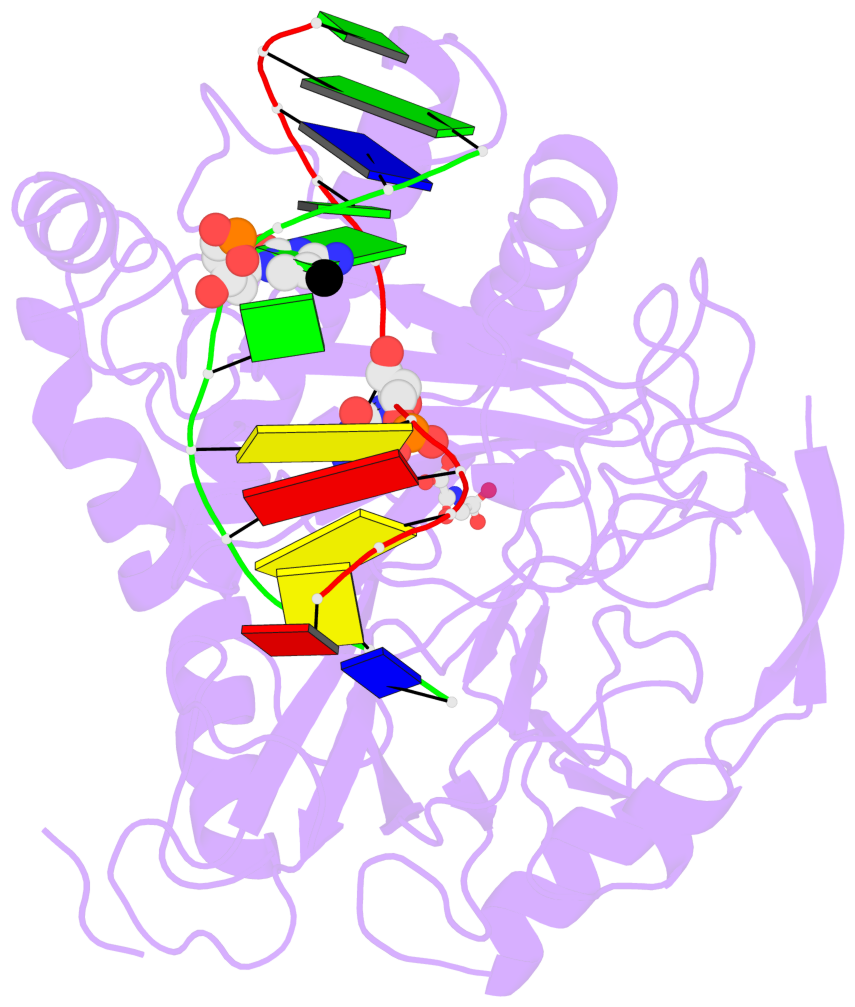
- The contacts include paired nucleotides (mostly a G in Watson-Crick G-C pairing), and
amino-acids within a 4.5-A distance cutoff to base atoms of 5mC.
- The structure is oriented in the base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
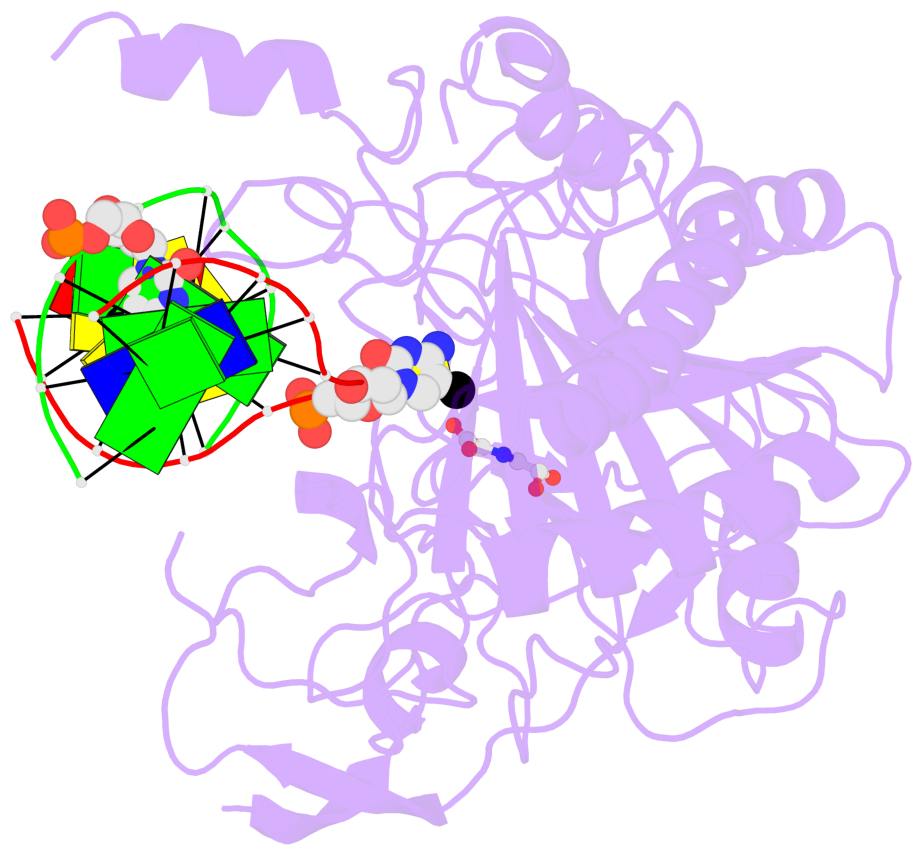
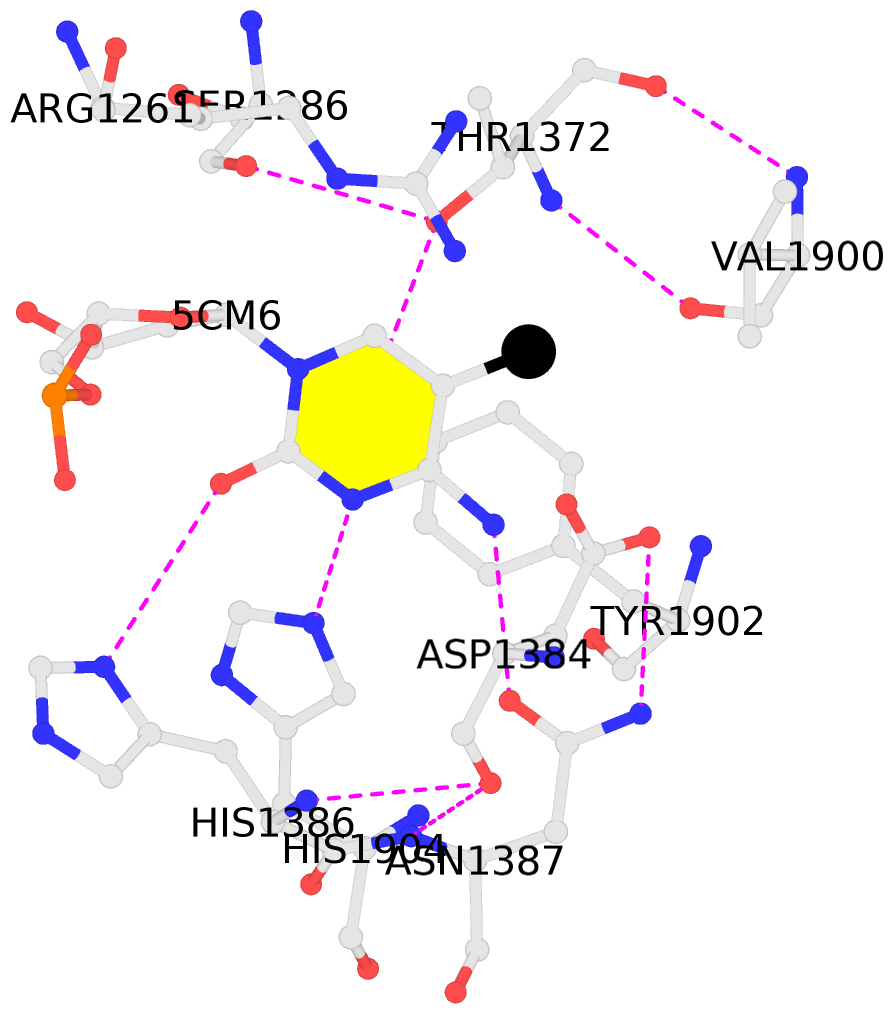
No. 1 B.5CM6: stacking-with-A.ARG1261 stacking-with-A.ASP1384 stacking-with-A.TYR1902 not-WC-paired not-in-duplex |
 |
|
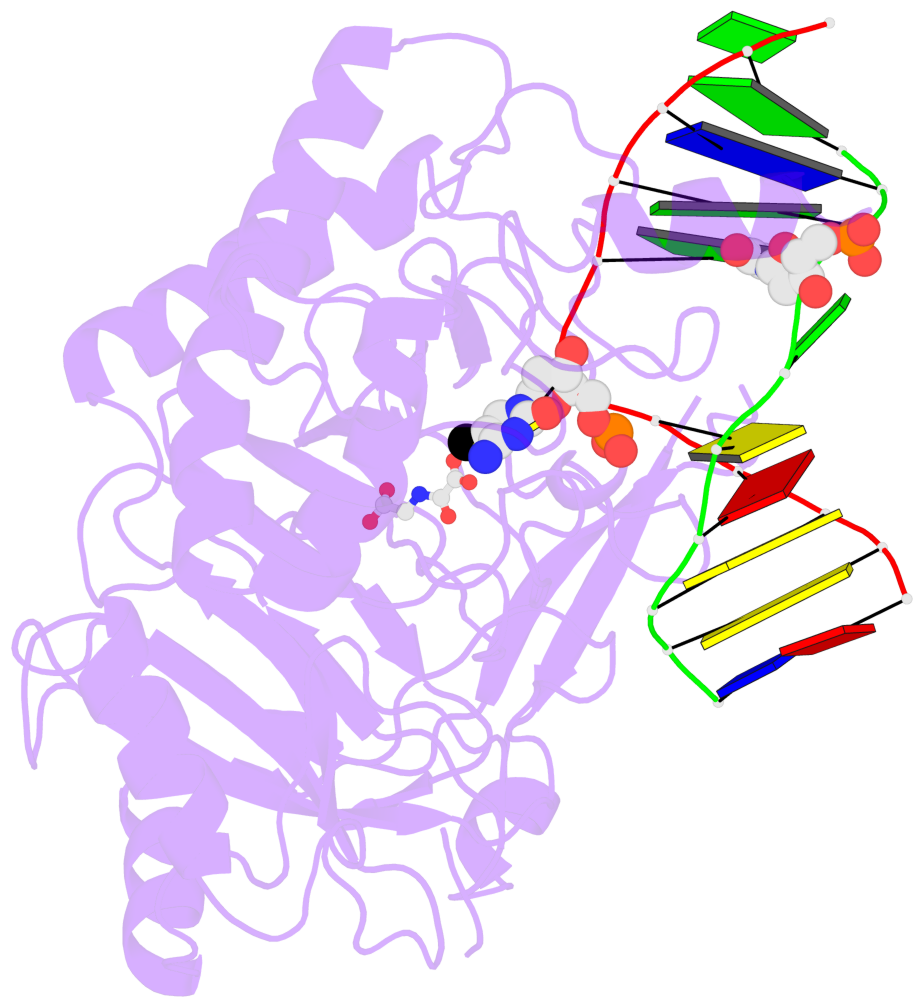
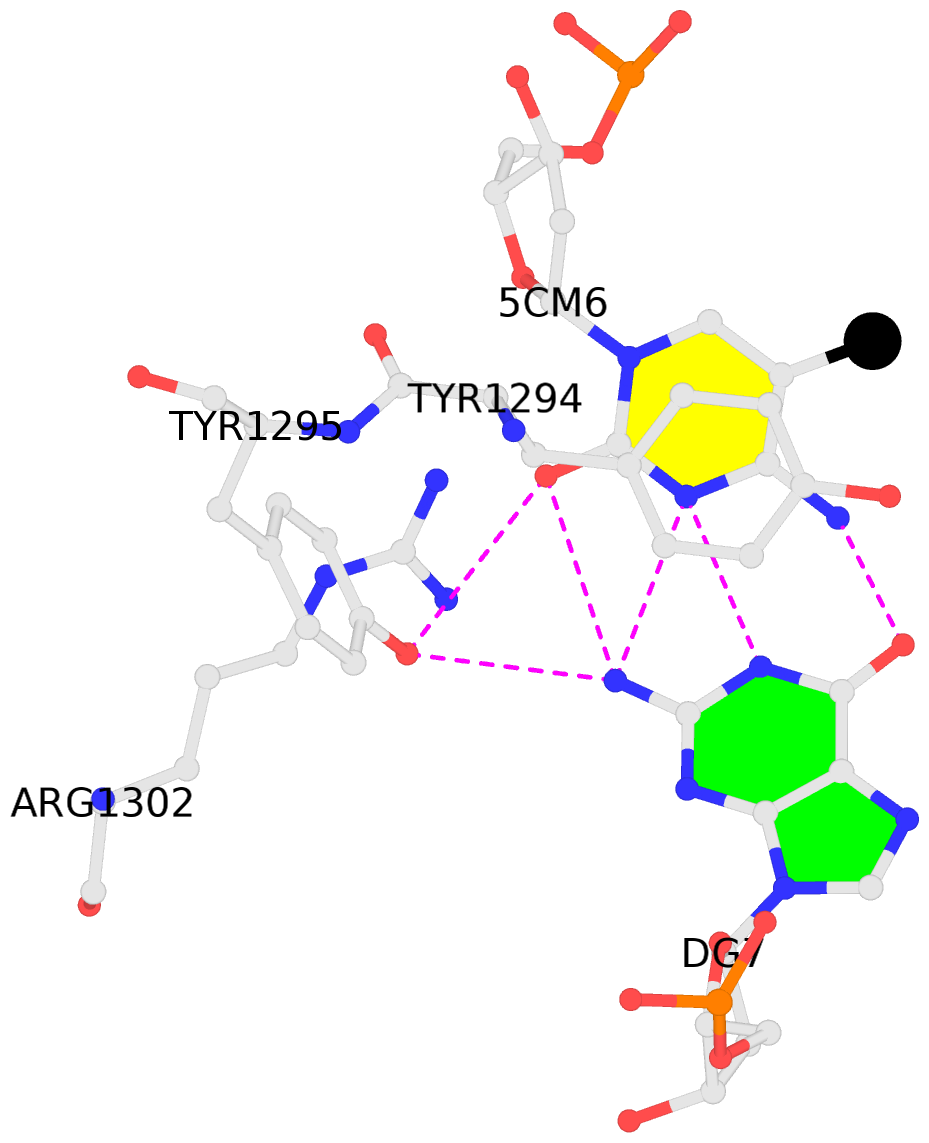
No. 2 C.5CM6: stacking-with-A.TYR1294 is-WC-paired is-in-duplex [-]:.GG/Cc. |
 |
|