Summary information and primary citation
- PDB-id
- 4pw7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-DNA
- Method
- X-ray (2.001 Å)
- Summary
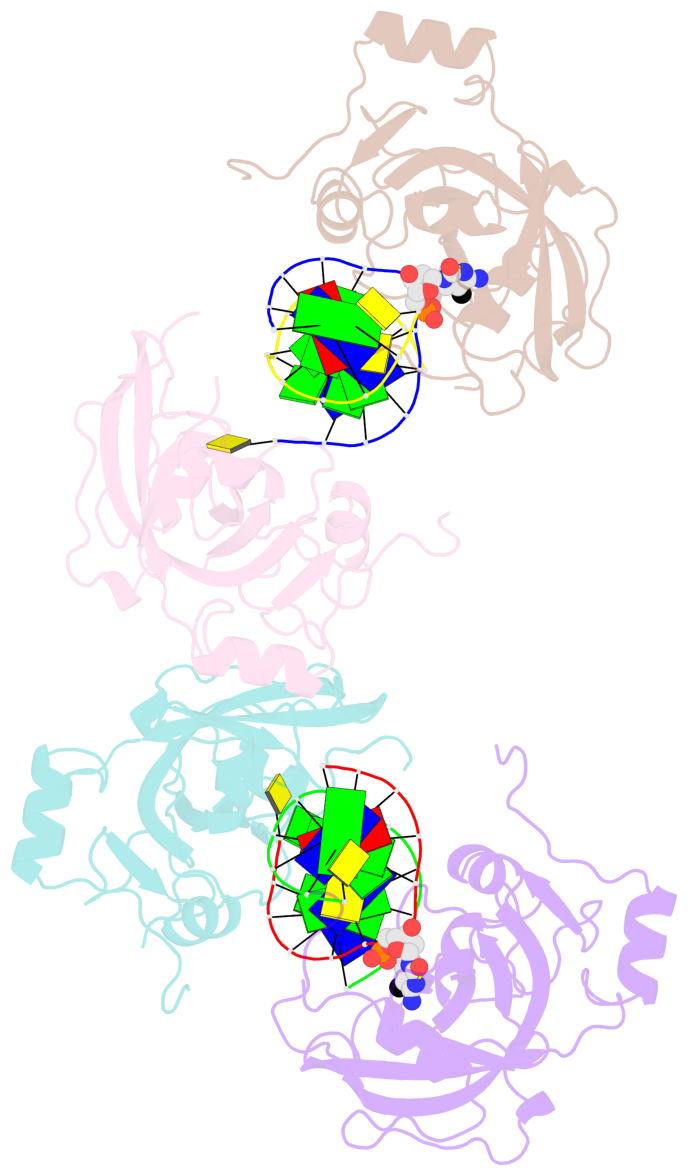
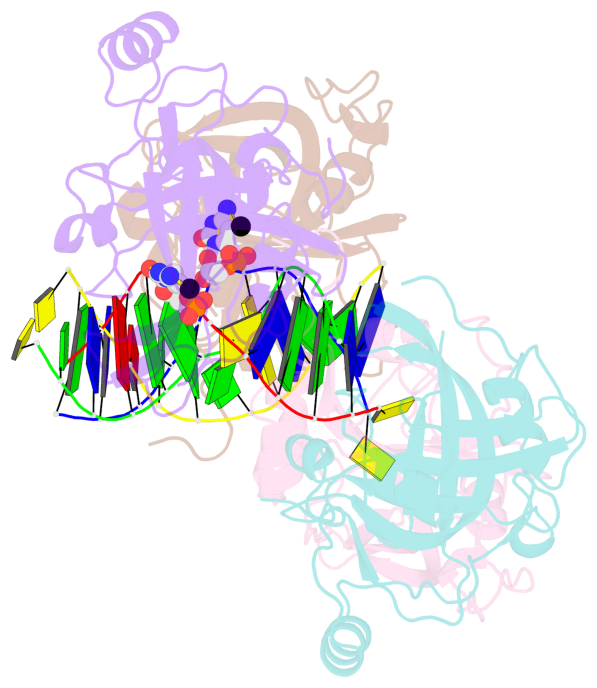
- Structure of uhrf2-sra in complex with a 5mc-containing DNA
- Reference
- Zhou T, Xiong J, Wang M, Yang N, Wong J, Zhu B, Xu RM (2014): "Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2." Mol.Cell, 54, 879-886. doi: 10.1016/j.molcel.2014.04.003.
- Abstract
- Methylated cytosine of CpG dinucleotides in vertebrates may be oxidized by Tet proteins, a process that can lead to DNA demethylation. The predominant oxidation product, 5-hydroxymethylcytosine (5hmC), has been implicated in embryogenesis, cell differentiation, and human diseases. Recently, the SRA domain of UHRF2 (UHRF2-SRA) has been reported to specifically recognize 5hmC, but how UHRF2 recognizes this modification is unclear. Here we report the structure of UHRF2-SRA in complex with a 5hmC-containing DNA. The structure reveals that the conformation of a phenylalanine allows the formation of an optimal 5hmC binding pocket, and a hydrogen bond between the hydroxyl group of 5hmC and UHRF2-SRA is critical for their preferential binding. Further structural and biochemical analyses unveiled the role of SRA domains as a versatile reader of modified DNA, and the knowledge should facilitate further understanding of the biological function of UHRF2 and the comprehension of DNA hydroxymethylation in general.
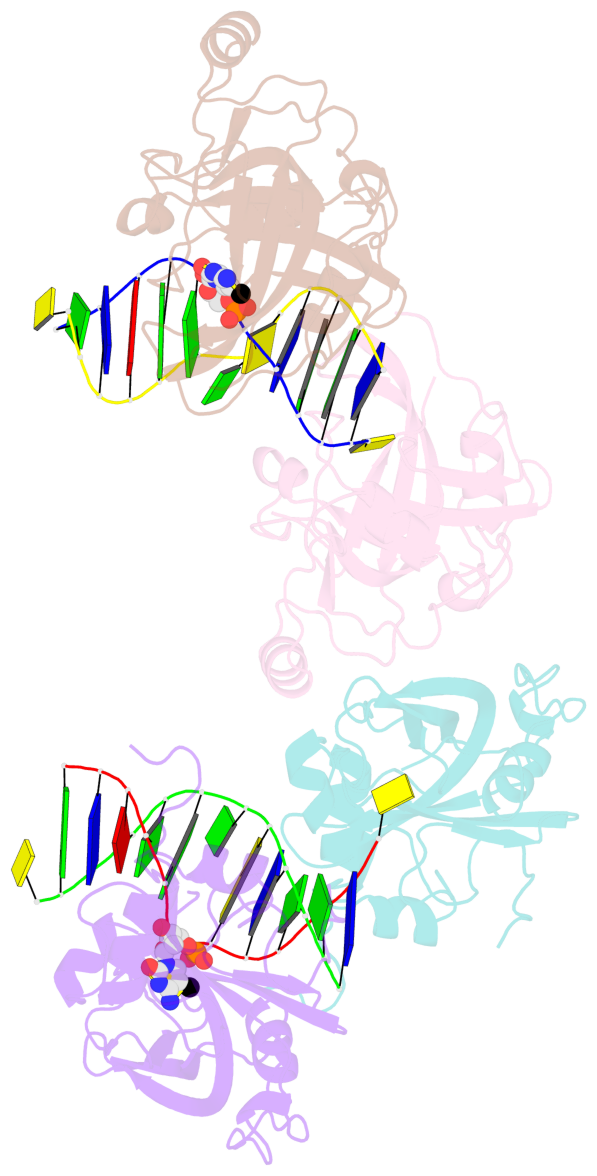
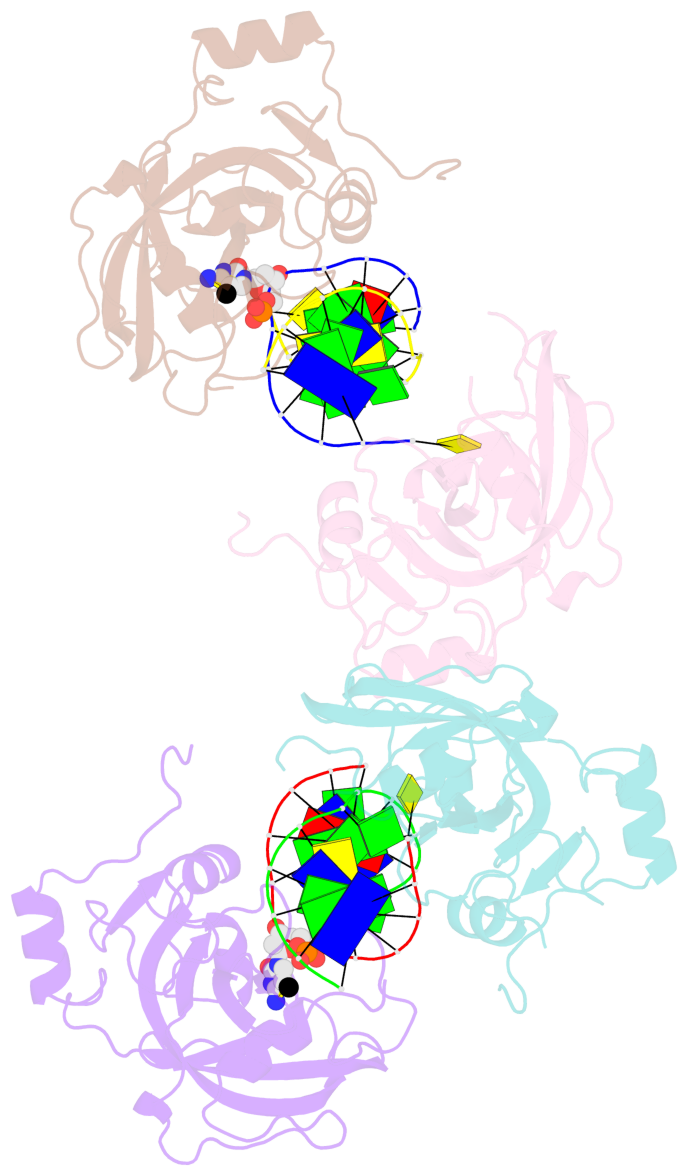
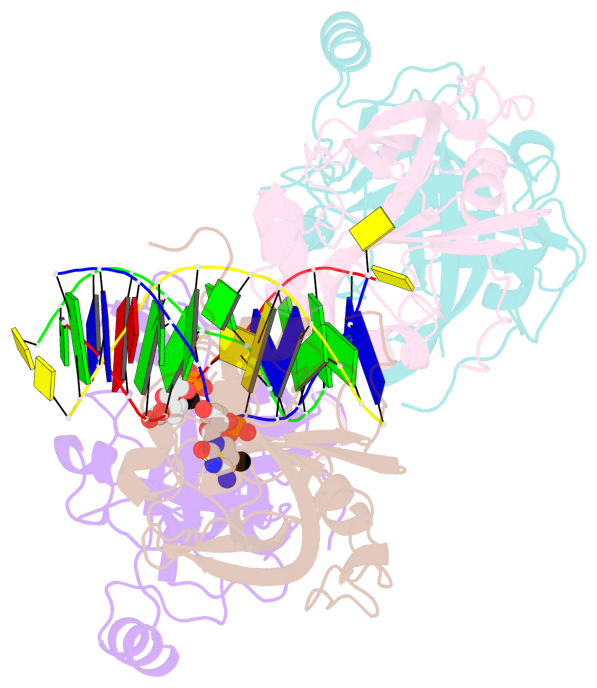
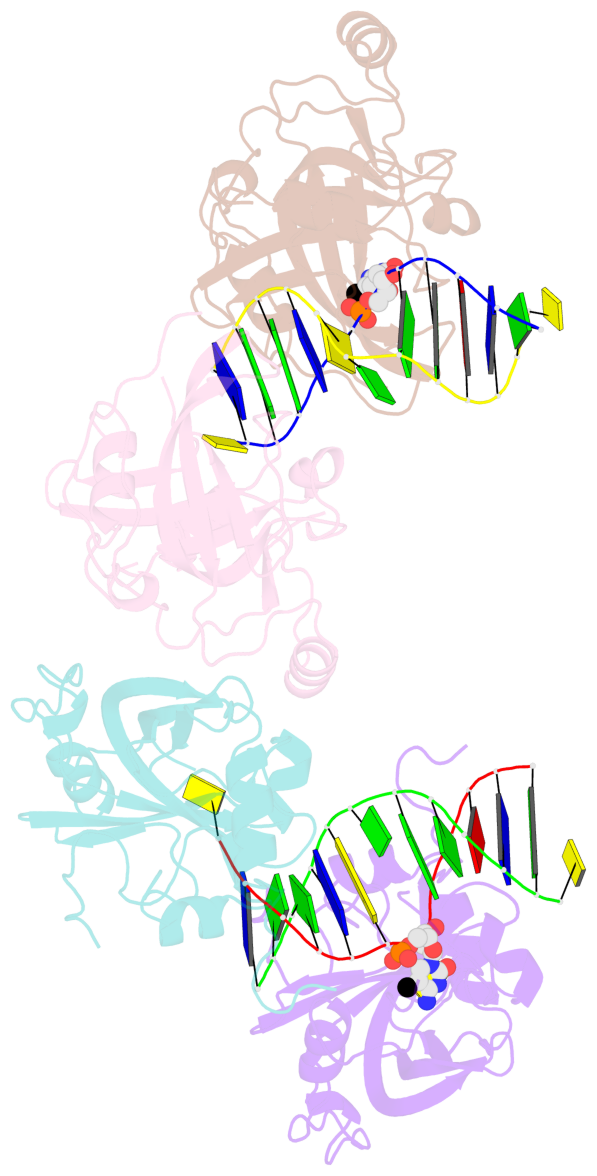
- The contacts include paired nucleotides (mostly a G in Watson-Crick G-C pairing), and
amino-acids within a 4.5-A distance cutoff to base atoms of 5mC.
- The structure is oriented in the base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
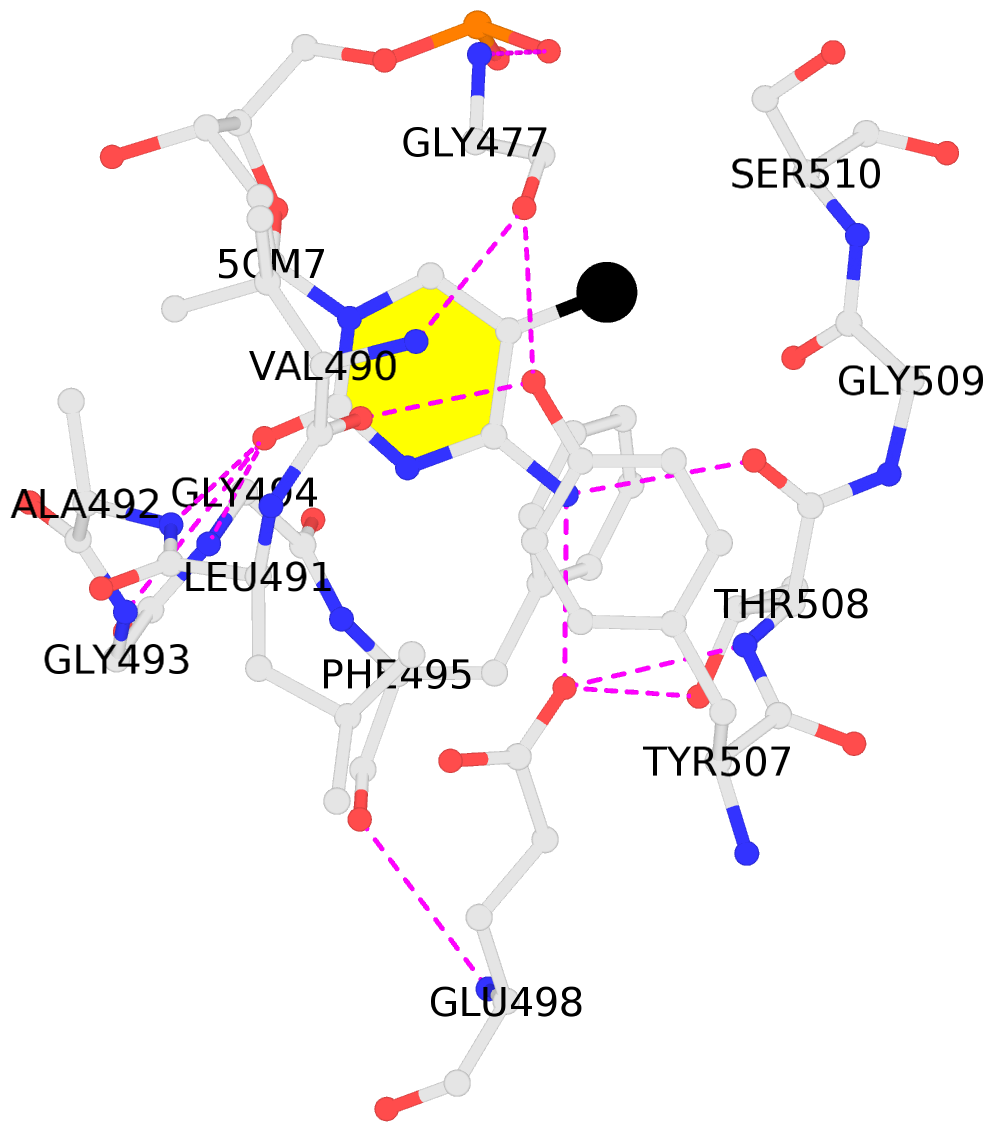
No. 1 C.5CM7: stacking-with-A.TYR507 not-WC-paired not-in-duplex |
 |
|
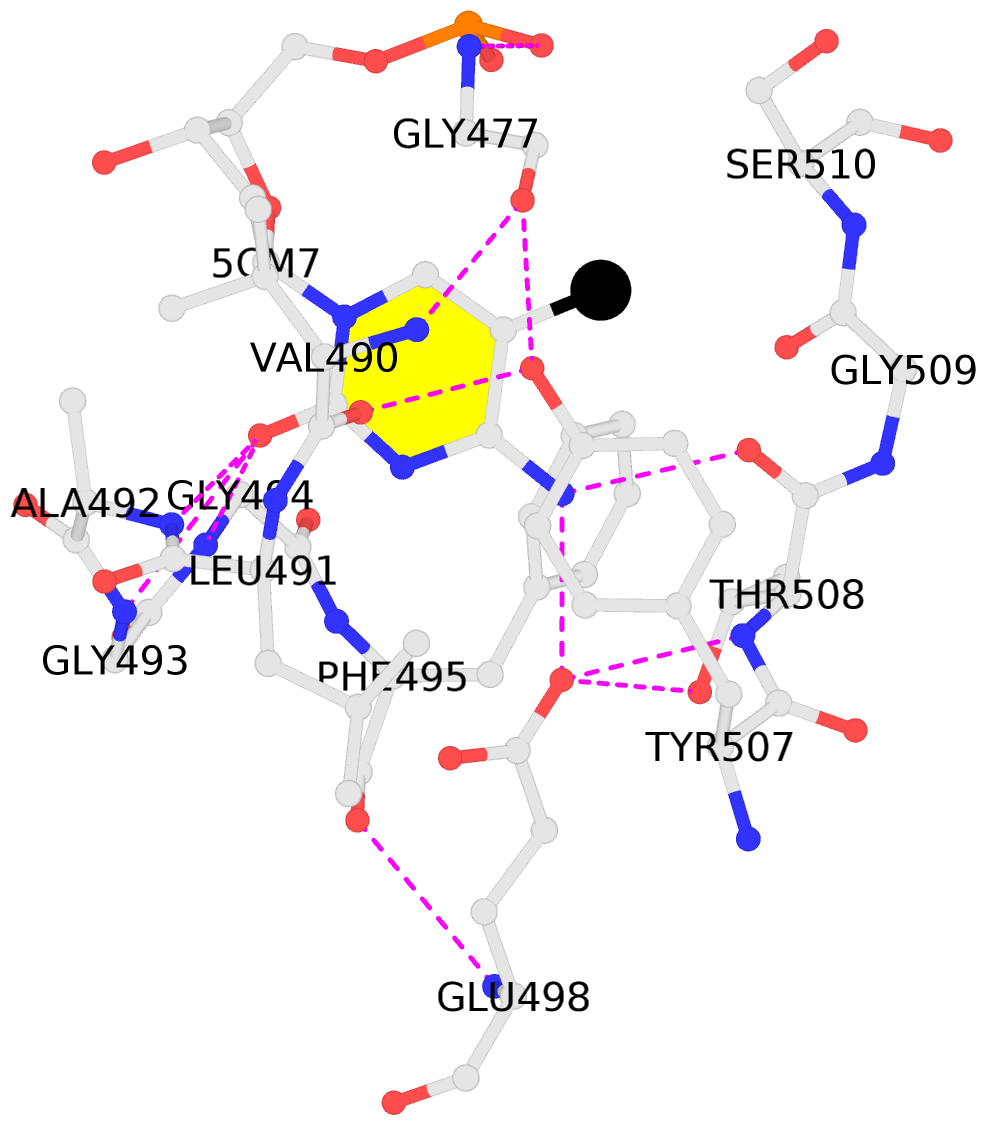
No. 2 G.5CM7: stacking-with-E.TYR507 not-WC-paired not-in-duplex |
 |
|