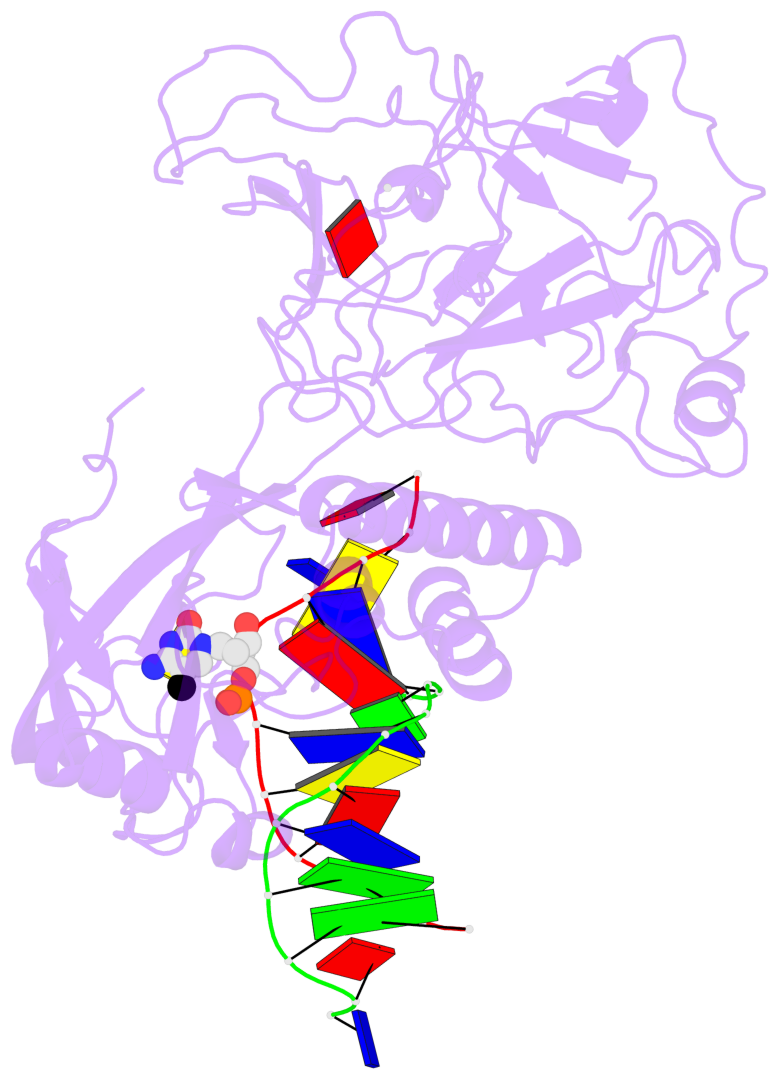
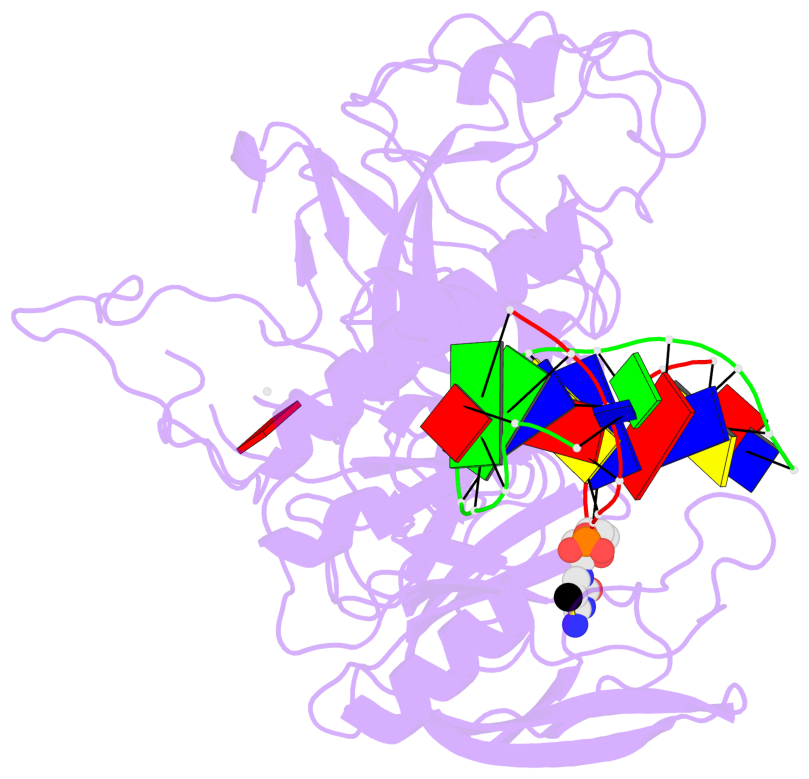
Summary information and primary citation
- PDB-id
- 4qen; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.002 Å)
- Summary
- Crystal structure of kryptonite in complex with mchh DNA and sah
- Reference
- Du J, Johnson LM, Groth M, Feng S, Hale CJ, Li S, Vashisht AA, Gallego-Bartolome J, Wohlschlegel JA, Patel DJ, Jacobsen SE (2014): "Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE." Mol.Cell, 55, 495-504. doi: 10.1016/j.molcel.2014.06.009.
- Abstract
- In Arabidopsis, CHG DNA methylation is controlled by the H3K9 methylation mark through a self-reinforcing loop between DNA methyltransferase CHROMOMETHYLASE3 (CMT3) and H3K9 histone methyltransferase KRYPTONITE/SUVH4 (KYP). We report on the structure of KYP in complex with methylated DNA, substrate H3 peptide, and cofactor SAH, thereby defining the spatial positioning of the SRA domain relative to the SET domain. The methylated DNA is bound by the SRA domain with the 5mC flipped out of the DNA, while the H3(1-15) peptide substrate binds between the SET and post-SET domains, with the ε-ammonium of K9 positioned adjacent to bound SAH. These structural insights, complemented by functional data on key mutants of residues lining the 5mC and H3K9-binding pockets within KYP, establish how methylated DNA recruits KYP to the histone substrate. Together, the structures of KYP and previously reported CMT3 complexes provide insights into molecular mechanisms linking DNA and histone methylation.
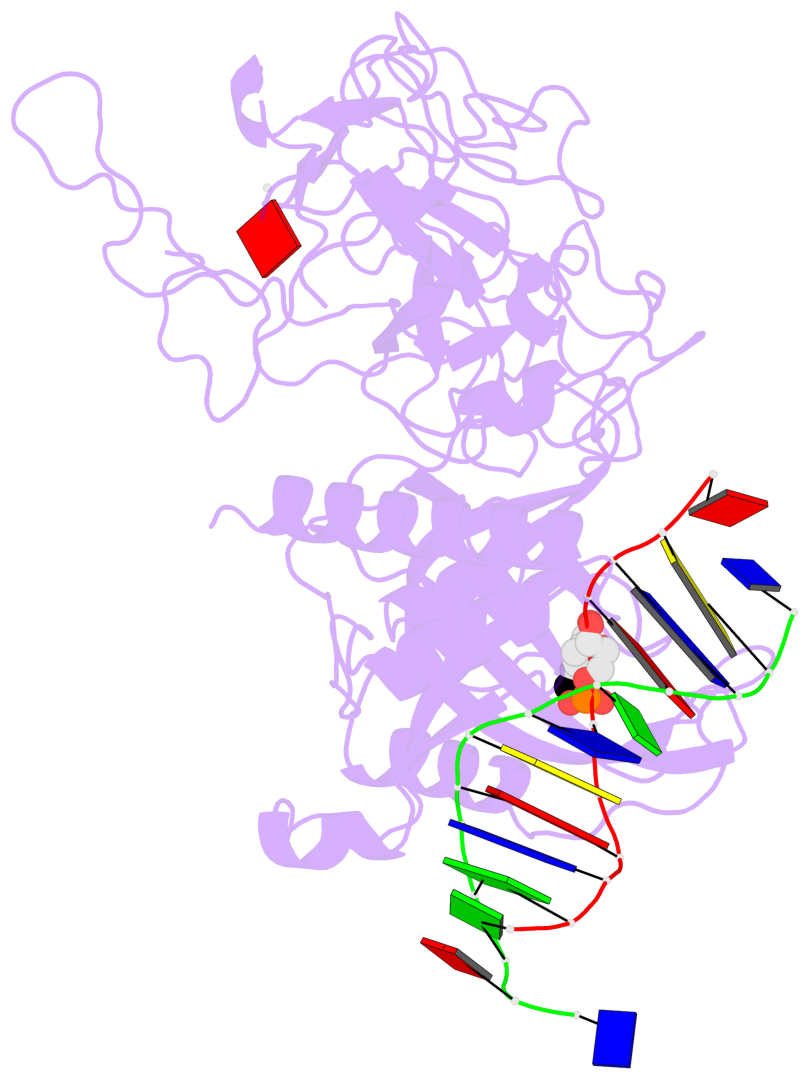
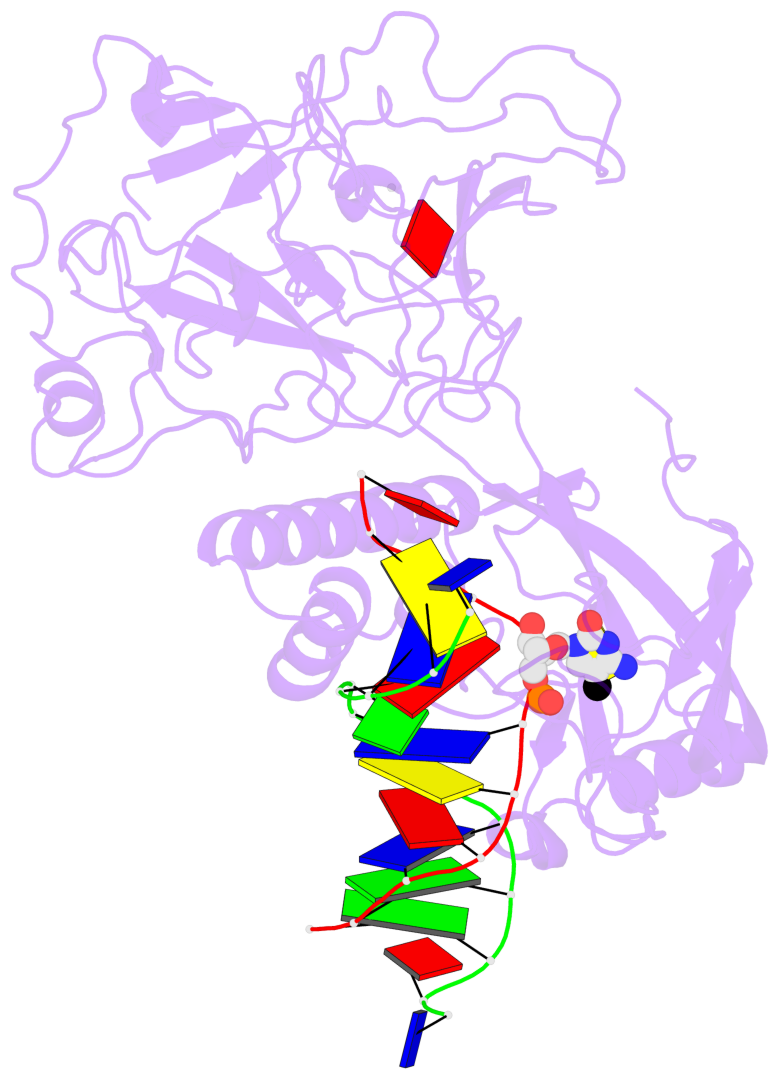
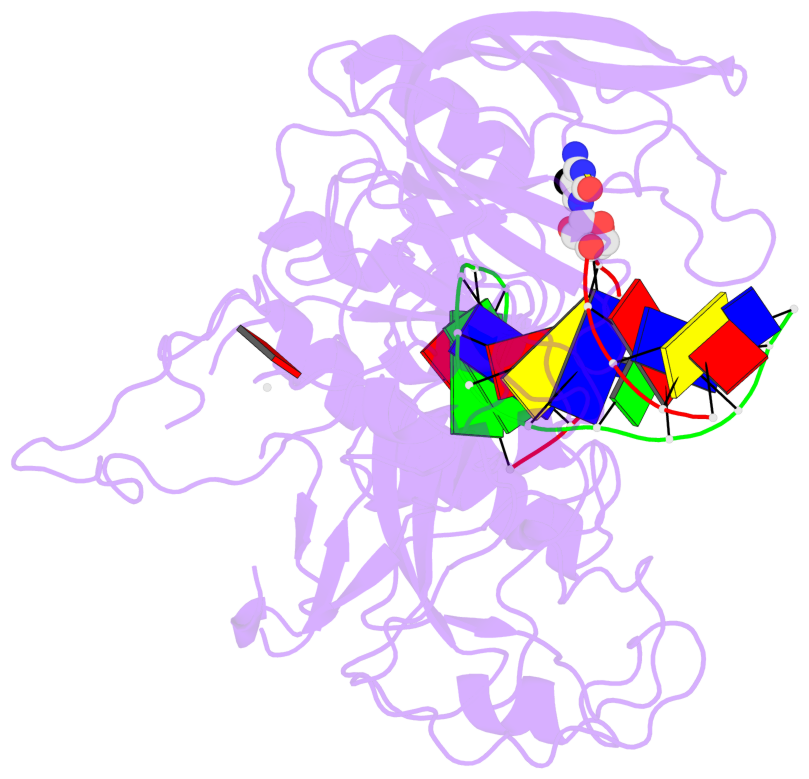
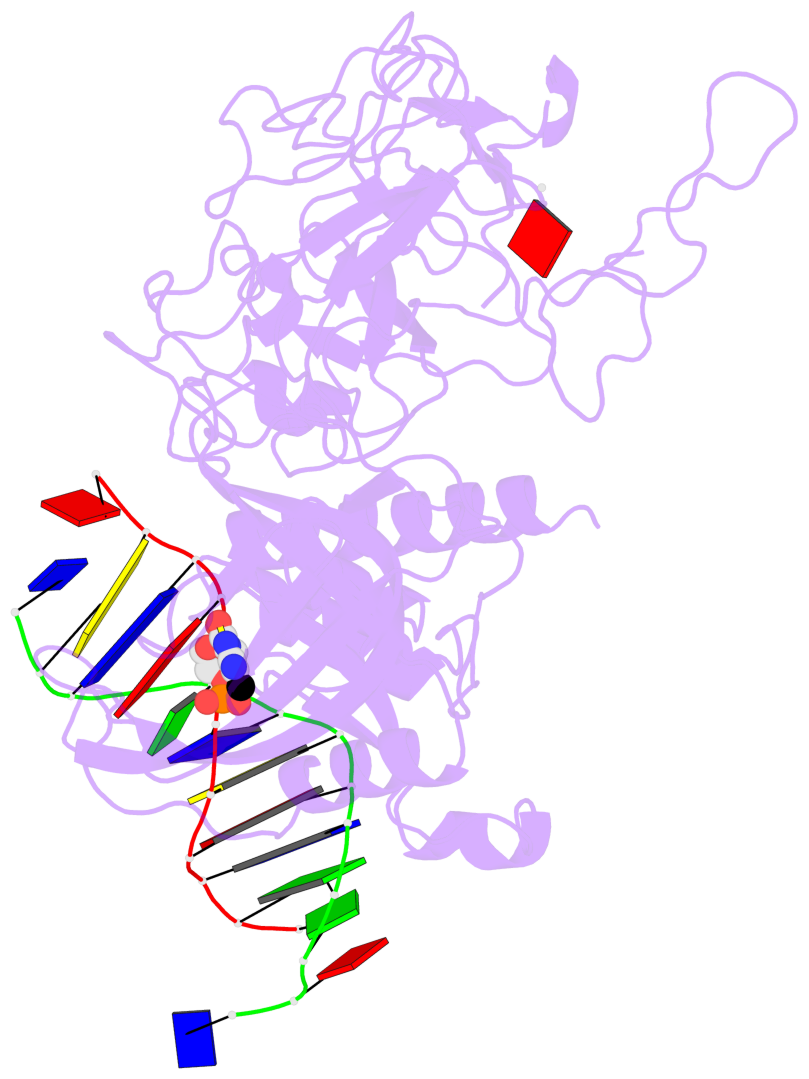
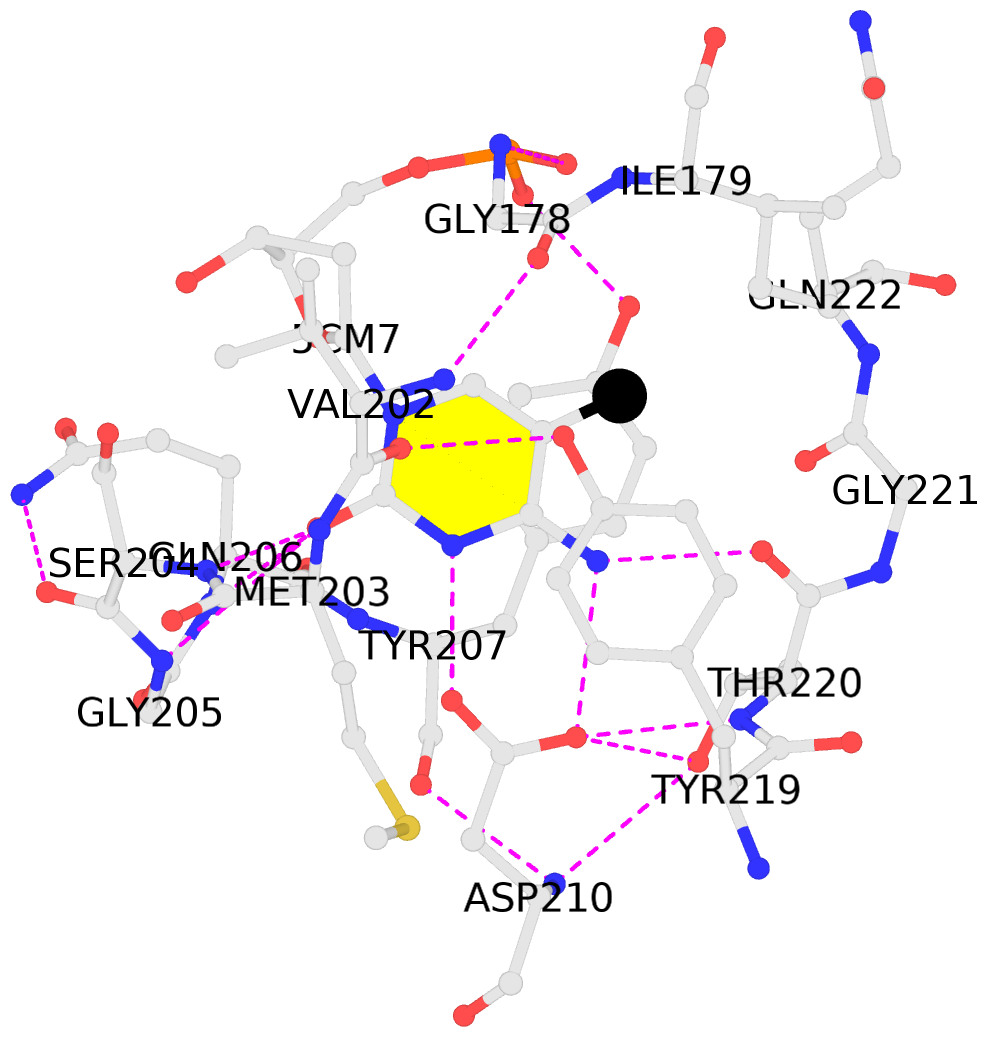
- The contacts include paired nucleotides (mostly a G in Watson-Crick G-C pairing), and
amino-acids within a 4.5-A distance cutoff to base atoms of 5mC.
- The structure is oriented in the base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
No. 1 C.5CM7: stacking-with-A.TYR207 stacking-with-A.TYR219 not-WC-paired not-in-duplex |
 |
|