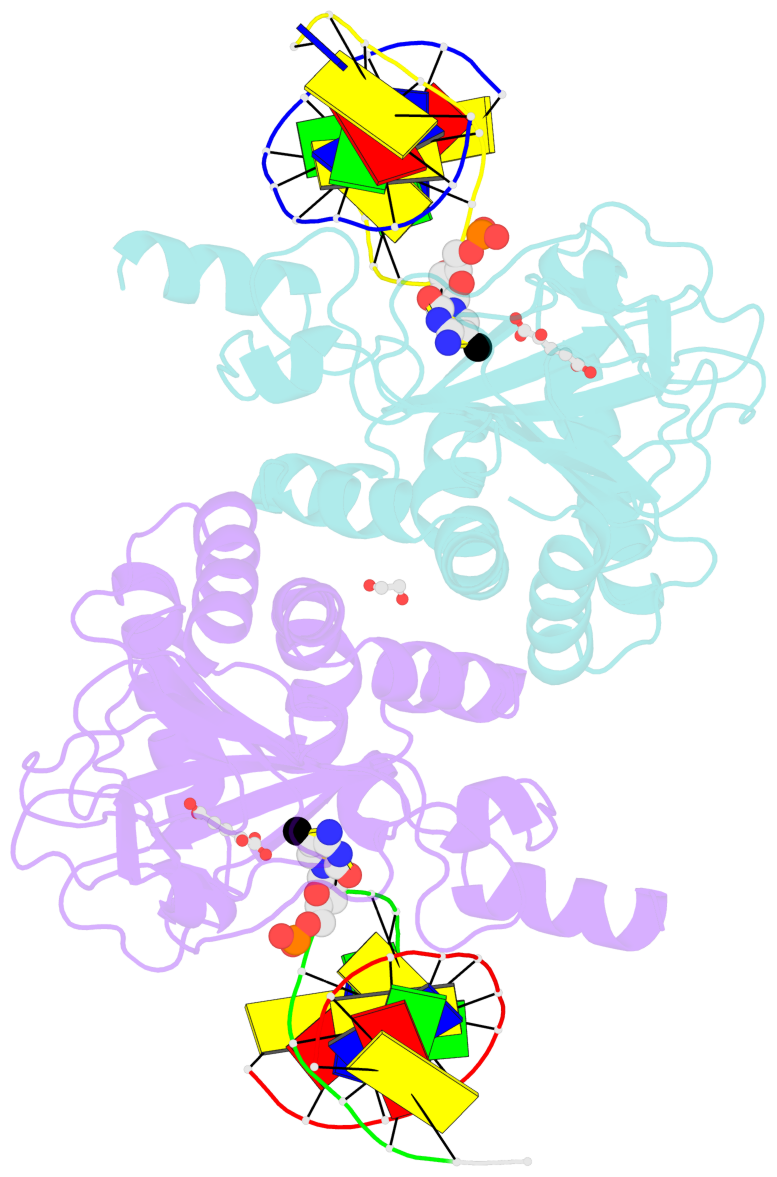
Summary information and primary citation
- PDB-id
- 5cg9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- oxidoreductase-DNA
- Method
- X-ray (2.693 Å)
- Summary
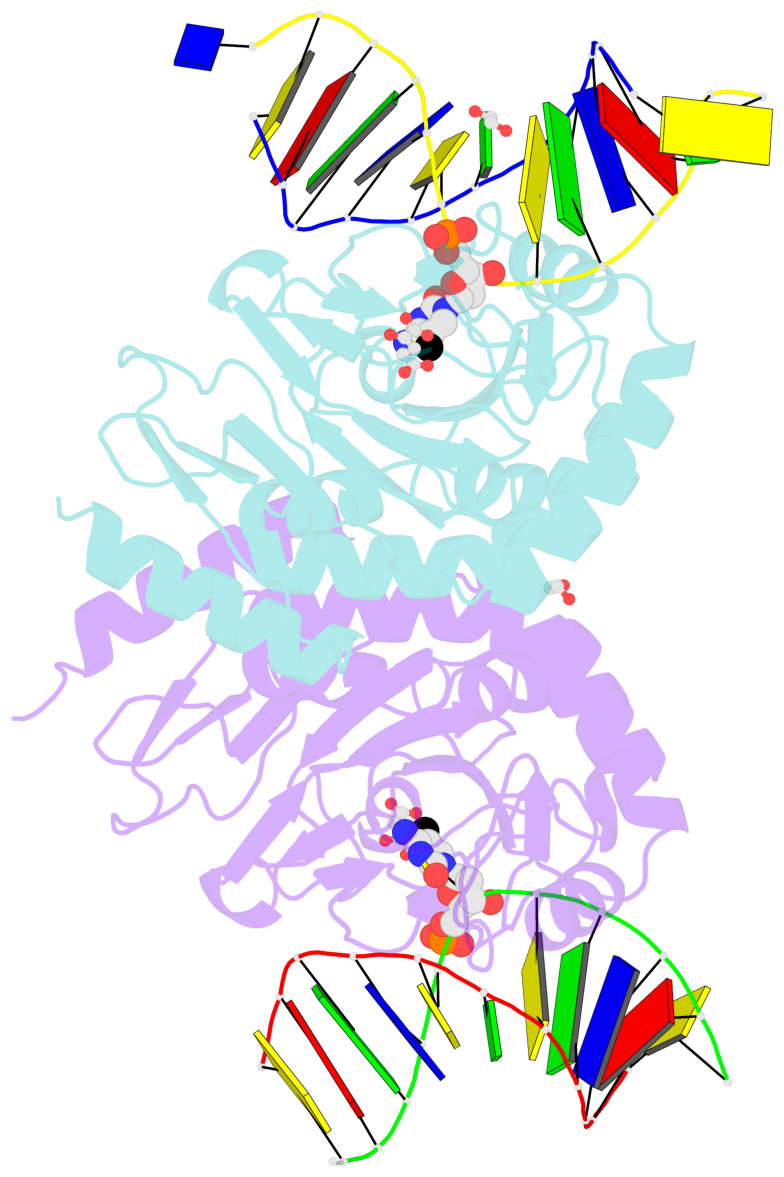
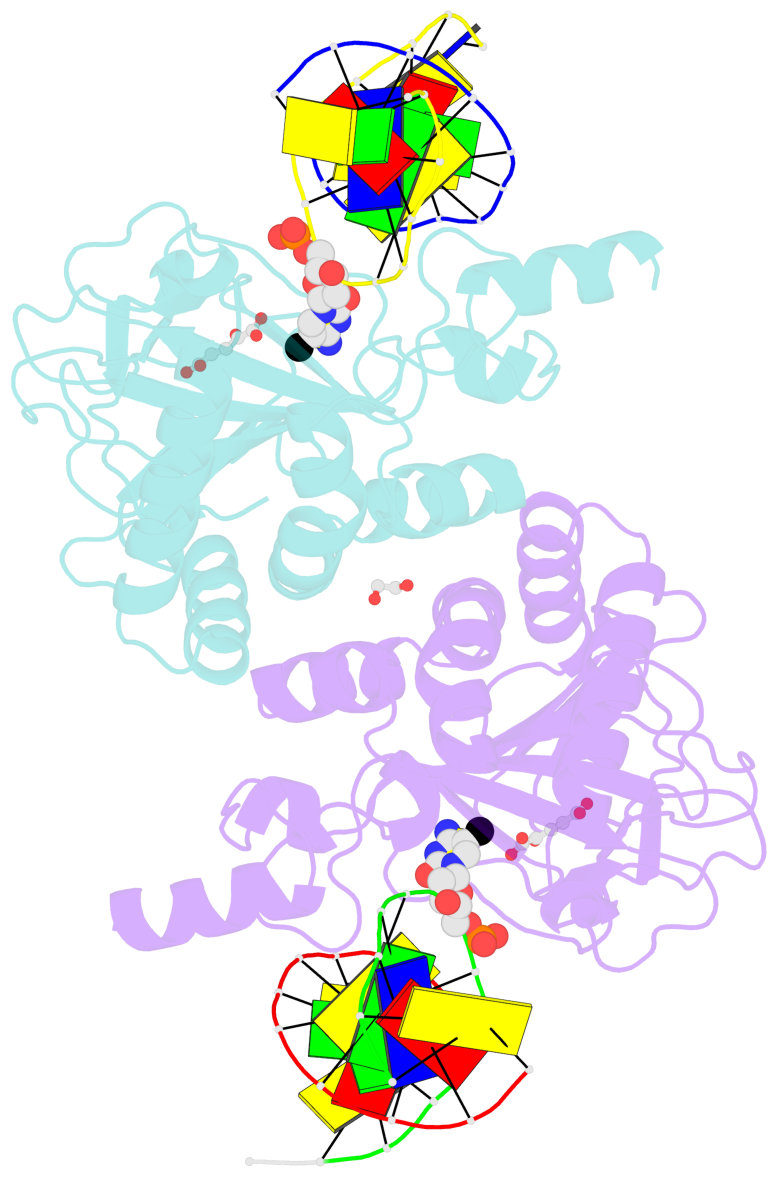
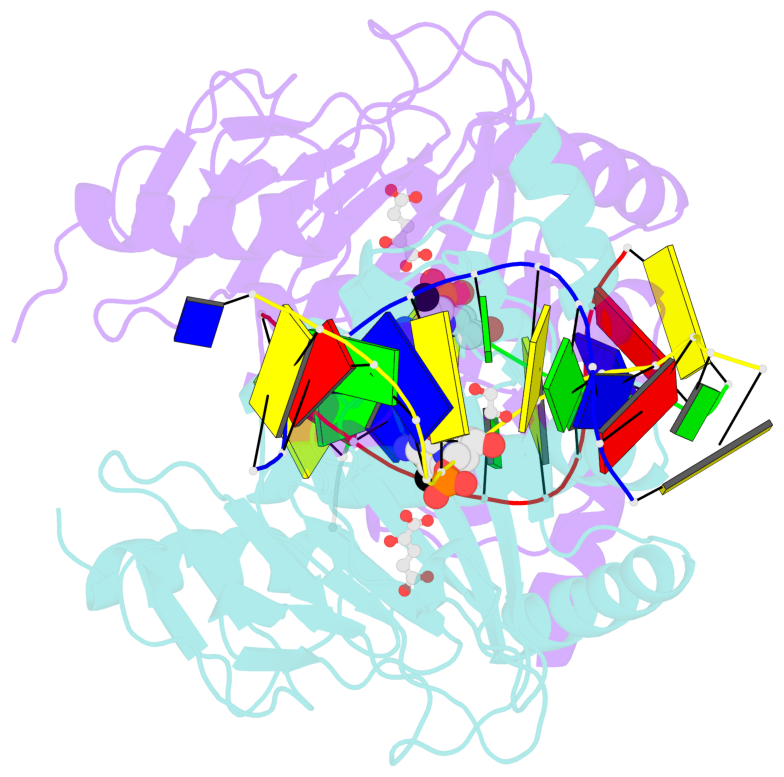
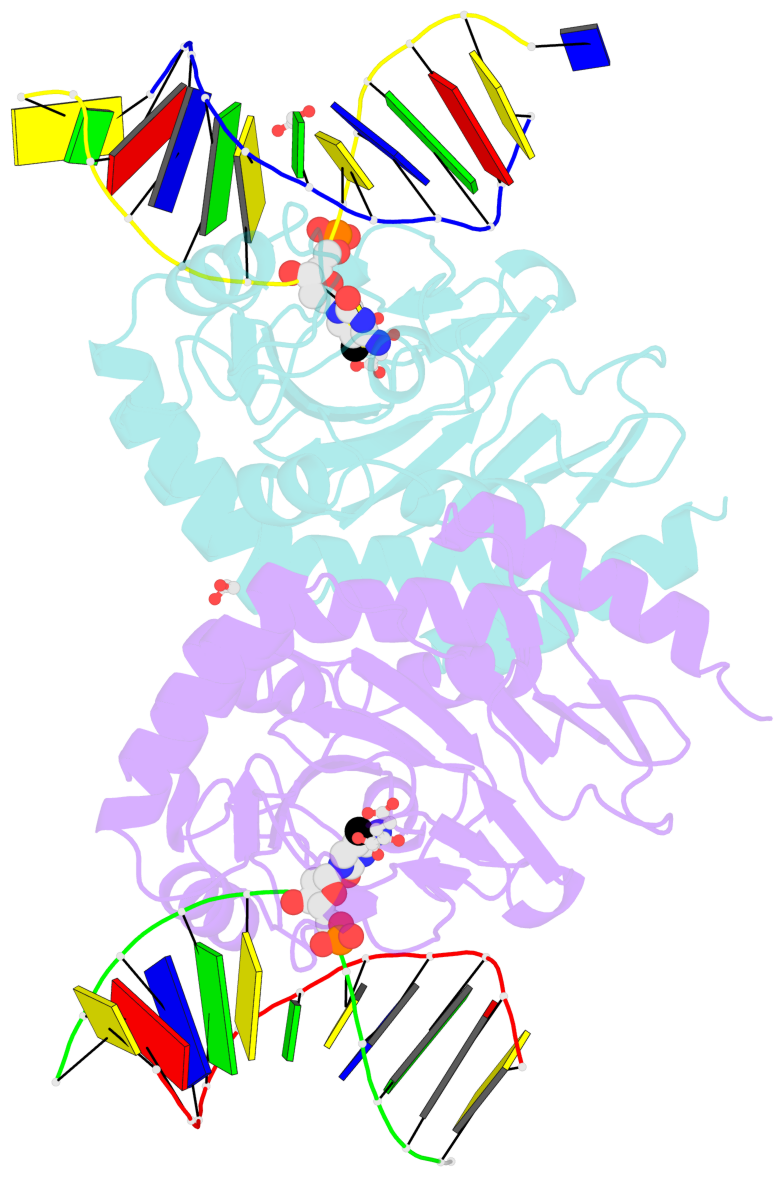
- Ngtet1 in complex with 5mc DNA in space group p3221
- Reference
- Hashimoto H, Pais JE, Dai N, Correa IR, Zhang X, Zheng Y, Cheng X (2015): "Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA." Nucleic Acids Res., 43, 10713-10721. doi: 10.1093/nar/gkv870.
- Abstract
- The family of ten-eleven translocation (Tet) dioxygenases is widely distributed across the eukaryotic tree of life, from mammals to the amoeboflagellate Naegleria gruberi. Like mammalian Tet proteins, the Naegleria Tet-like protein, NgTet1, acts on 5-methylcytosine (5mC) and generates 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in three consecutive, Fe(II)- and α-ketoglutarate-dependent oxidation reactions. The two intermediates, 5hmC and 5fC, could be considered either as the reaction product of the previous enzymatic cycle or the substrate for the next cycle. Here we present a new crystal structure of NgTet1 in complex with DNA containing a 5hmC. Along with the previously solved NgTet1-5mC structure, the two complexes offer a detailed picture of the active site at individual stages of the reaction cycle. In the crystal, the hydroxymethyl (OH-CH2-) moiety of 5hmC points to the metal center, representing the reaction product of 5mC hydroxylation. The hydroxyl oxygen atom could be rotated away from the metal center, to a hydrophobic pocket formed by Ala212, Val293 and Phe295. Such rotation turns the hydroxyl oxygen atom away from the product conformation, and exposes the target CH2 towards the metal-ligand water molecule, where a dioxygen O2 molecule would occupy to initiate the next round of reaction by abstracting a hydrogen atom from the substrate. The Ala212-to-Val (A212V) mutant profoundly limits the product to 5hmC, probably because the reduced hydrophobic pocket size restricts the binding of 5hmC as a substrate.
- The contacts include paired nucleotides (mostly a G in Watson-Crick G-C pairing), and
amino-acids within a 4.5-A distance cutoff to base atoms of 5mC.
- The structure is oriented in the base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
No. 1 C.5CM20: stacking-with-A.ARG224 stacking-with-A.ASP231 stacking-with-A.PHE295 not-WC-paired not-in-duplex |
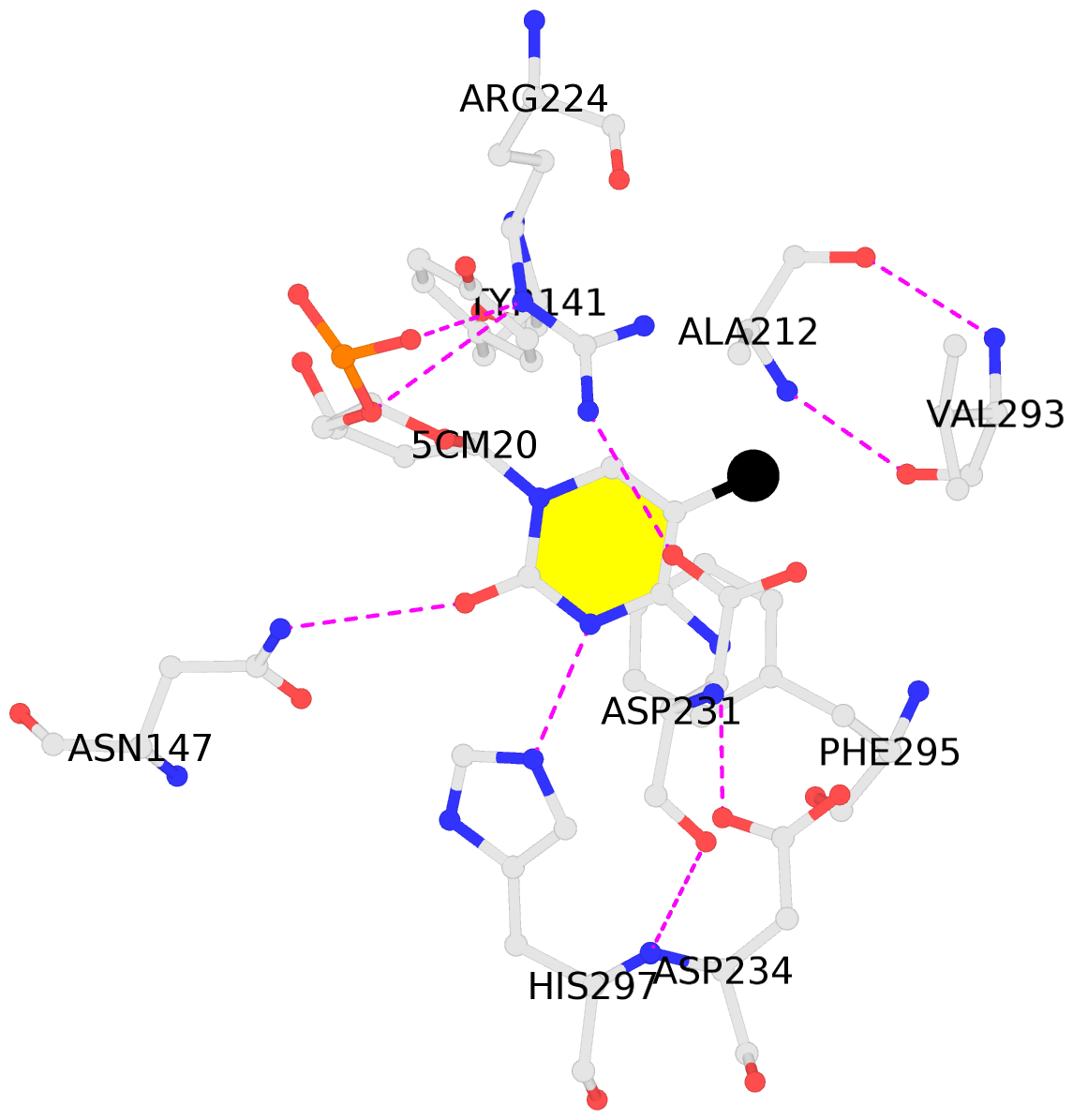 |
|
No. 2 F.5CM20: stacking-with-D.ARG224 stacking-with-D.ASP231 stacking-with-D.PHE295 not-WC-paired not-in-duplex |
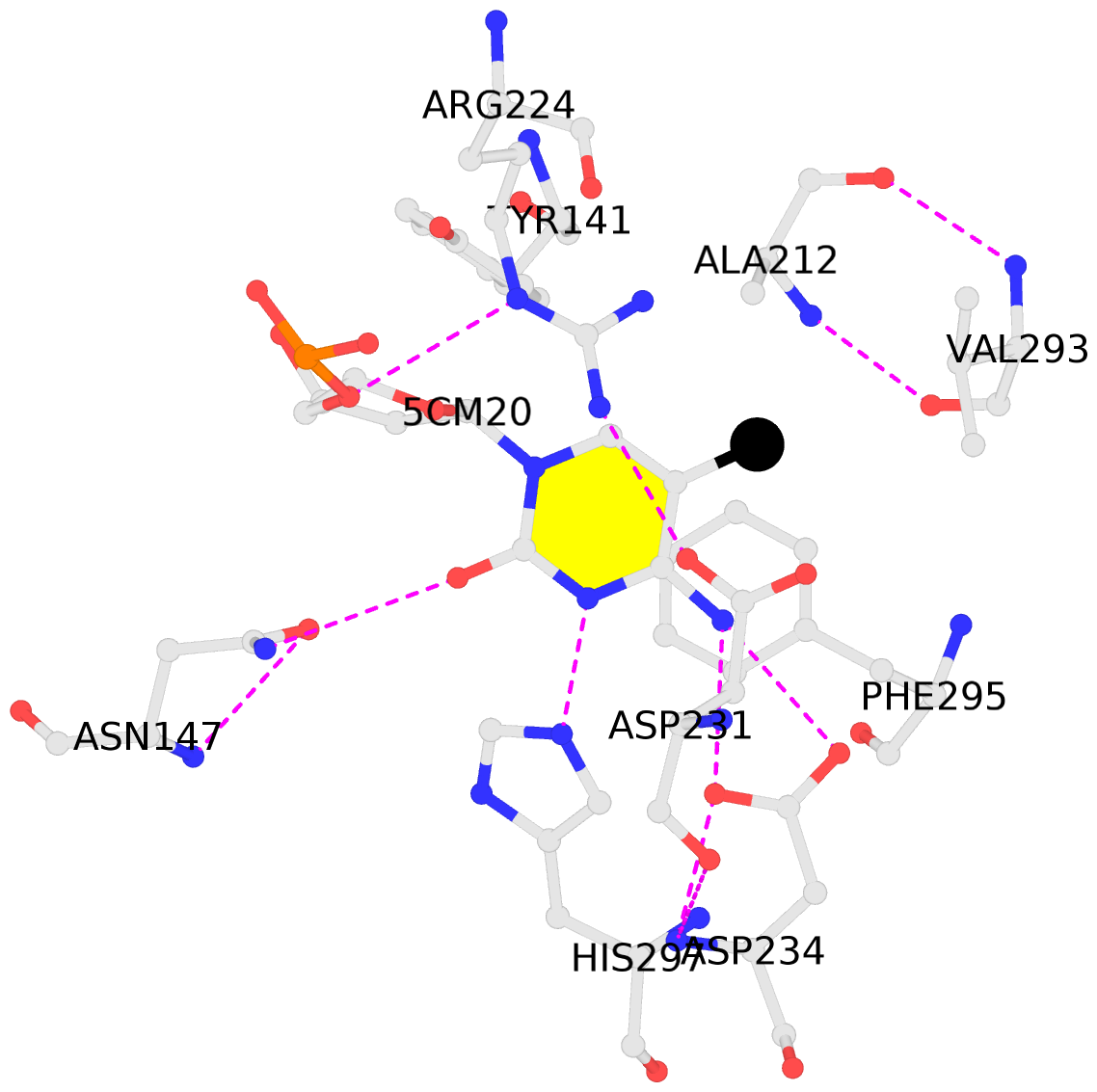 |
|