Summary information and primary citation
- PDB-id
- 6ykf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (1.48 Å)
- Summary
- Vcam4i restriction endonuclease in the presence of 5mc-modified ssDNA
- Reference
- Pastor M, Czapinska H, Helbrecht I, Krakowska K, Lutz T, Xu SY, Bochtler M (2021): "Crystal structures of the EVE-HNH endonuclease VcaM4I in the presence and absence of DNA." Nucleic Acids Res., 49, 1708-1723. doi: 10.1093/nar/gkaa1218.
- Abstract
- Many modification-dependent restriction endonucleases (MDREs) are fusions of a PUA superfamily modification sensor domain and a nuclease catalytic domain. EVE domains belong to the PUA superfamily, and are present in MDREs in combination with HNH nuclease domains. Here, we present a biochemical characterization of the EVE-HNH endonuclease VcaM4I and crystal structures of the protein alone, with EVE domain bound to either 5mC modified dsDNA or to 5mC/5hmC containing ssDNA. The EVE domain is moderately specific for 5mC/5hmC containing DNA according to EMSA experiments. It flips the modified nucleotide, to accommodate it in a hydrophobic pocket of the enzyme, primarily formed by P24, W82 and Y130 residues. In the crystallized conformation, the EVE domain and linker helix between the two domains block DNA binding to the catalytic domain. Removal of the EVE domain and inter-domain linker, but not of the EVE domain alone converts VcaM4I into a non-specific toxic nuclease. The role of the key residues in the EVE and HNH domains of VcaM4I is confirmed by digestion and restriction assays with the enzyme variants that differ from the wild-type by changes to the base binding pocket or to the catalytic residues.
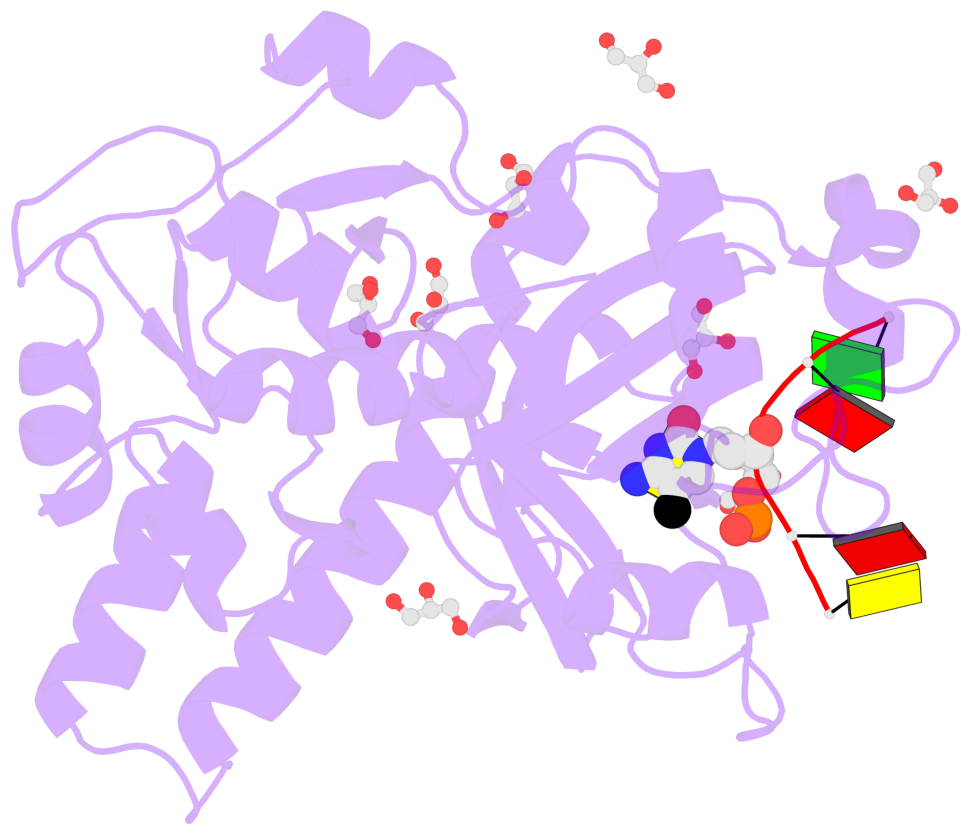
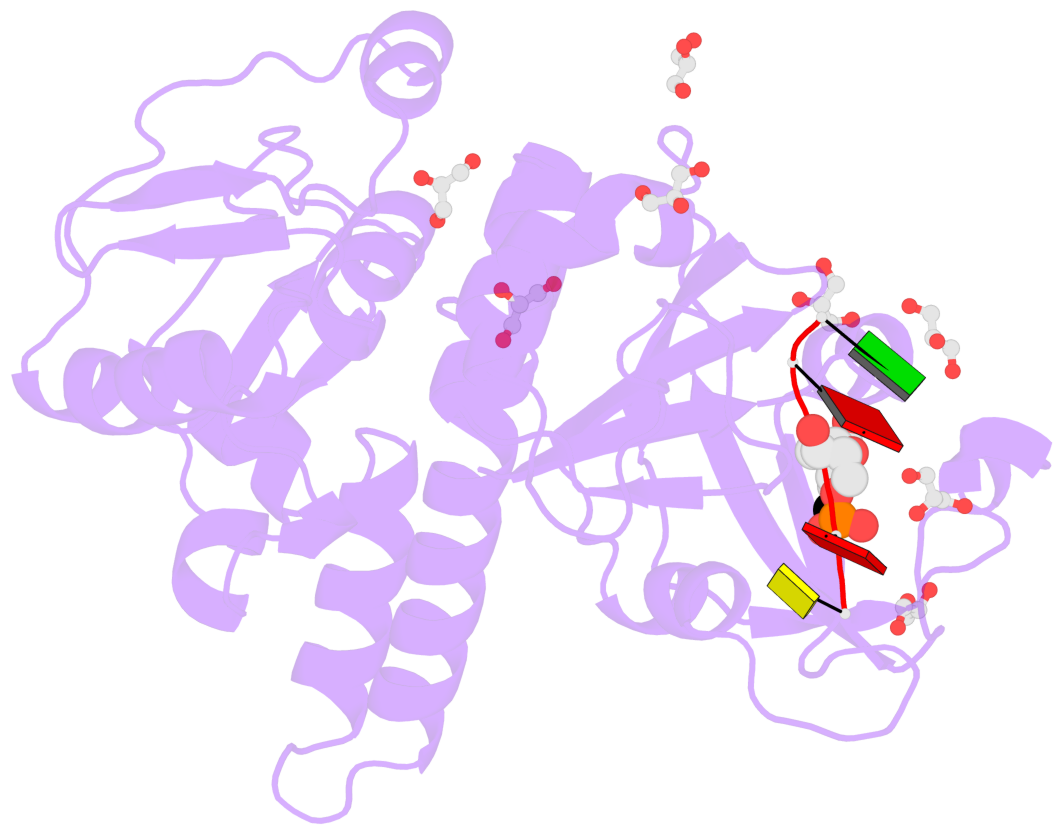
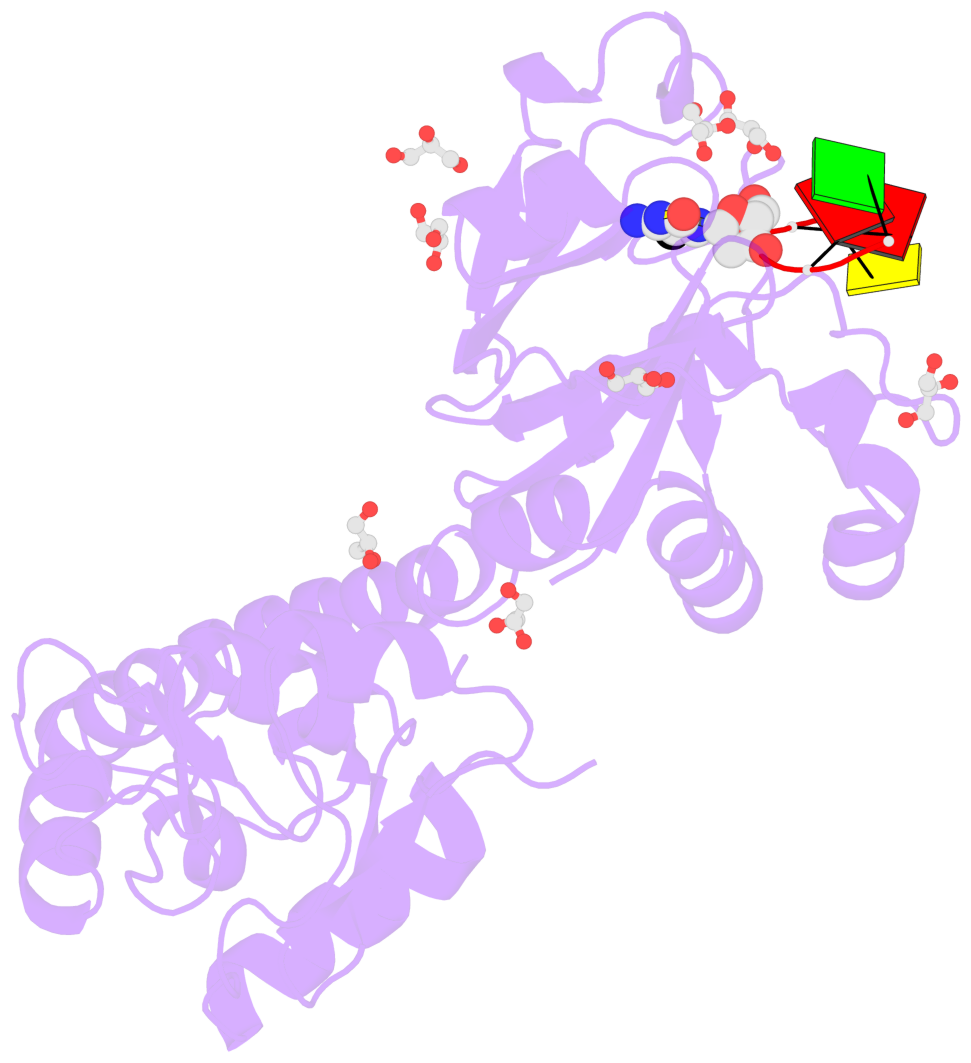
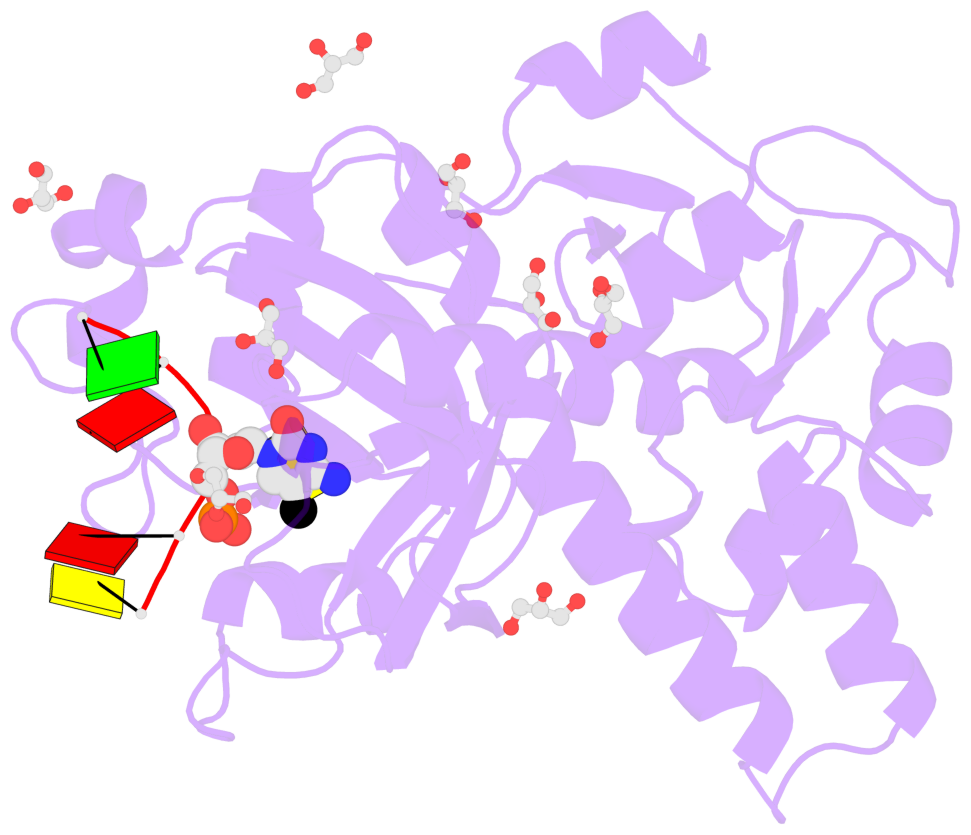
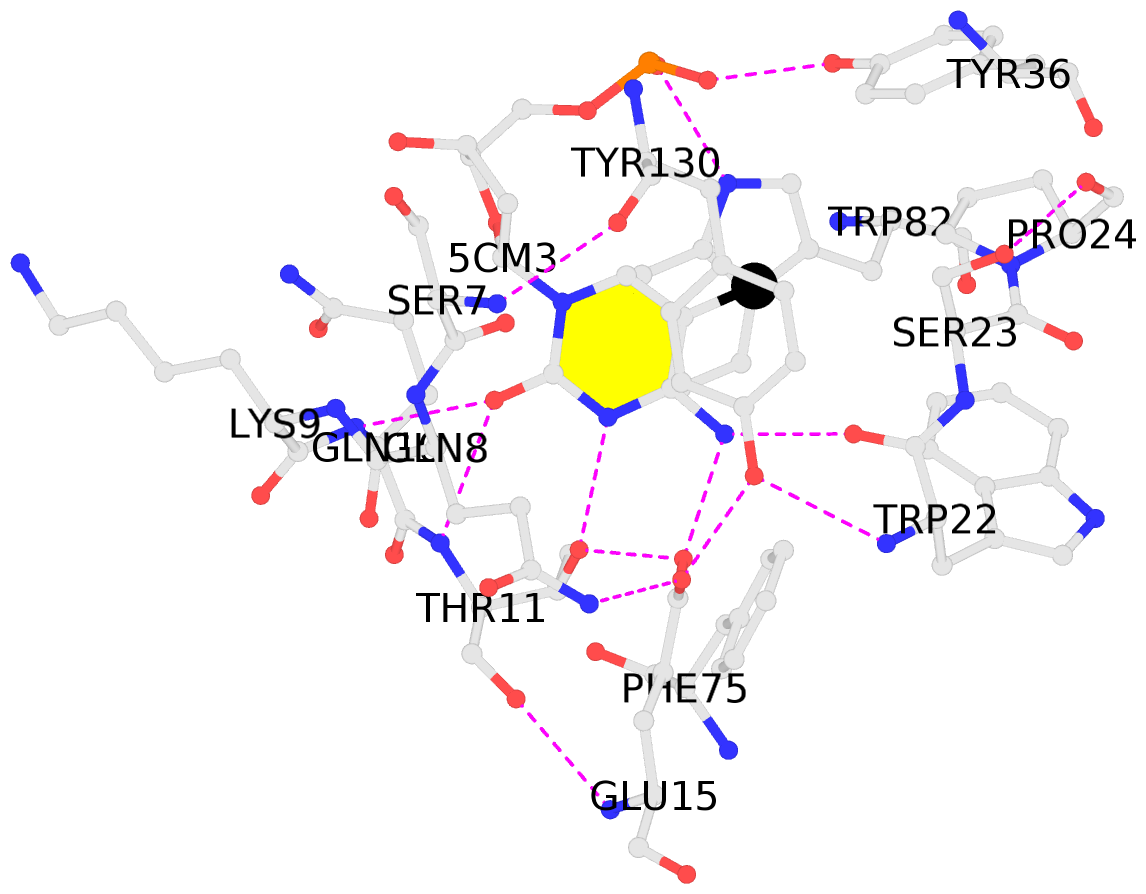
- The contacts include paired nucleotides (mostly a G in Watson-Crick G-C pairing), and
amino-acids within a 4.5-A distance cutoff to base atoms of 5mC.
- The structure is oriented in the base reference frame of 5mC, allowing for easy comparison
and direct superimposition between entries.
- The black sphere (•) denotes the 5-methyl carbon atom in 5mC.
No. 1 B.5CM3: stacking-with-A.TRP82 stacking-with-A.TYR130 not-WC-paired not-in-duplex |
 |
|