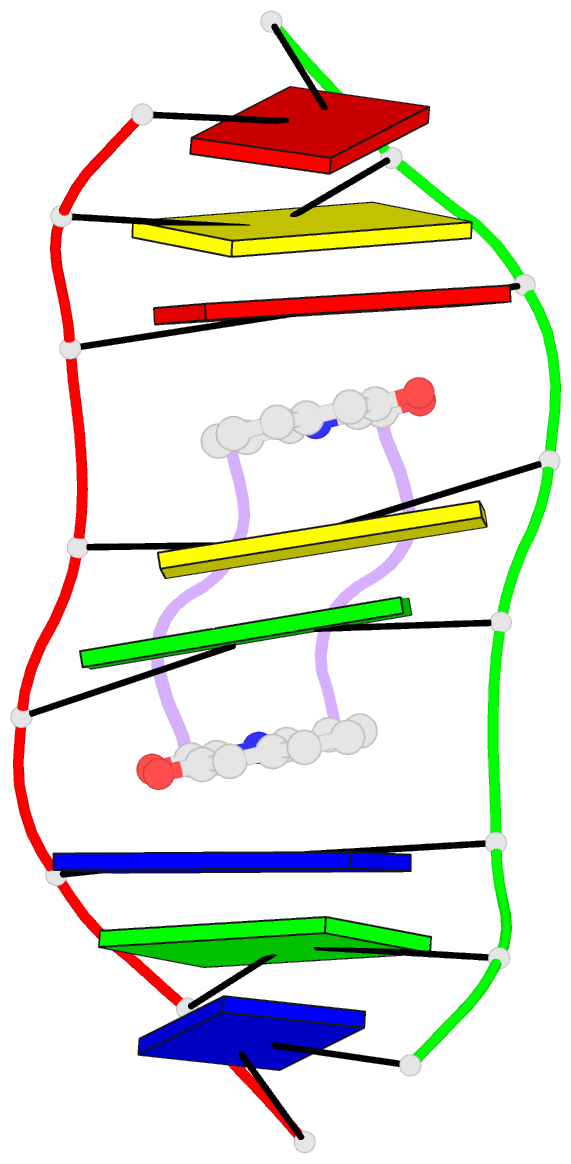
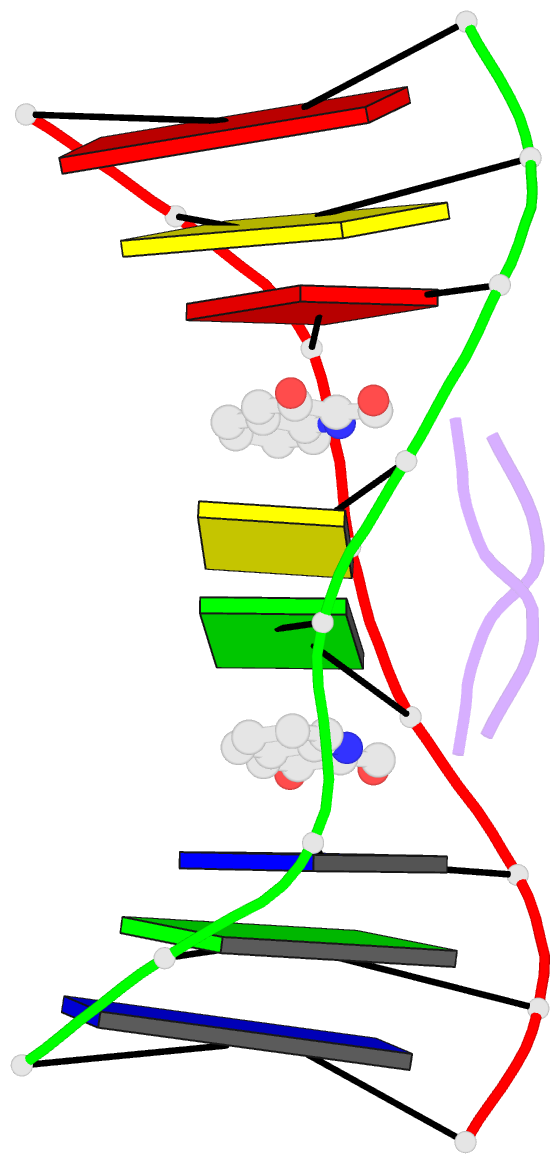
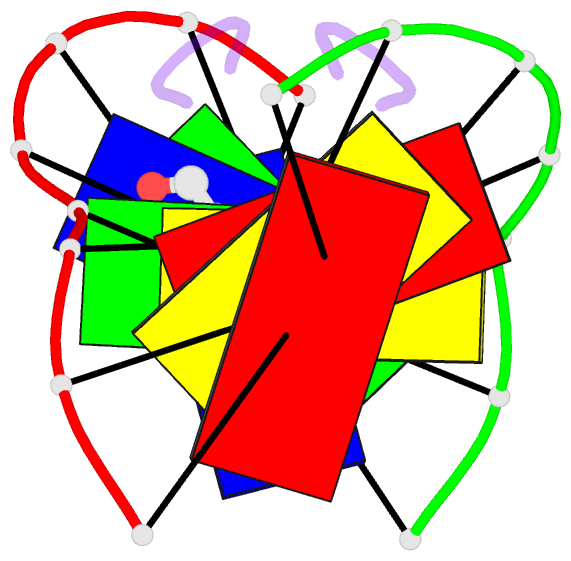
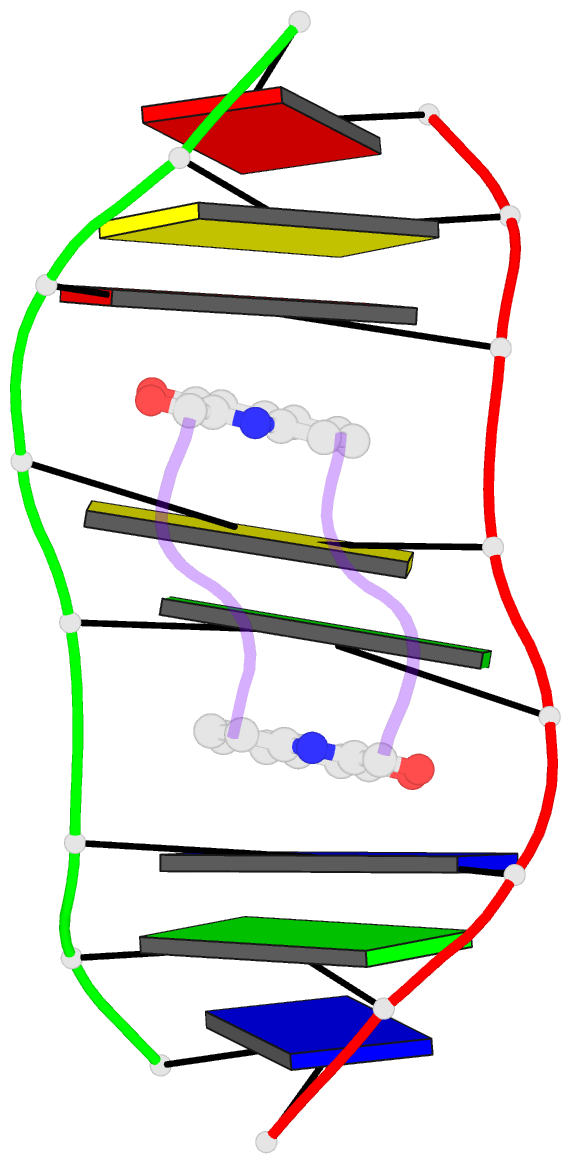
Summary information and primary citation
- PDB-id
- 193d; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA-antibiotic
- Method
- NMR
- Summary
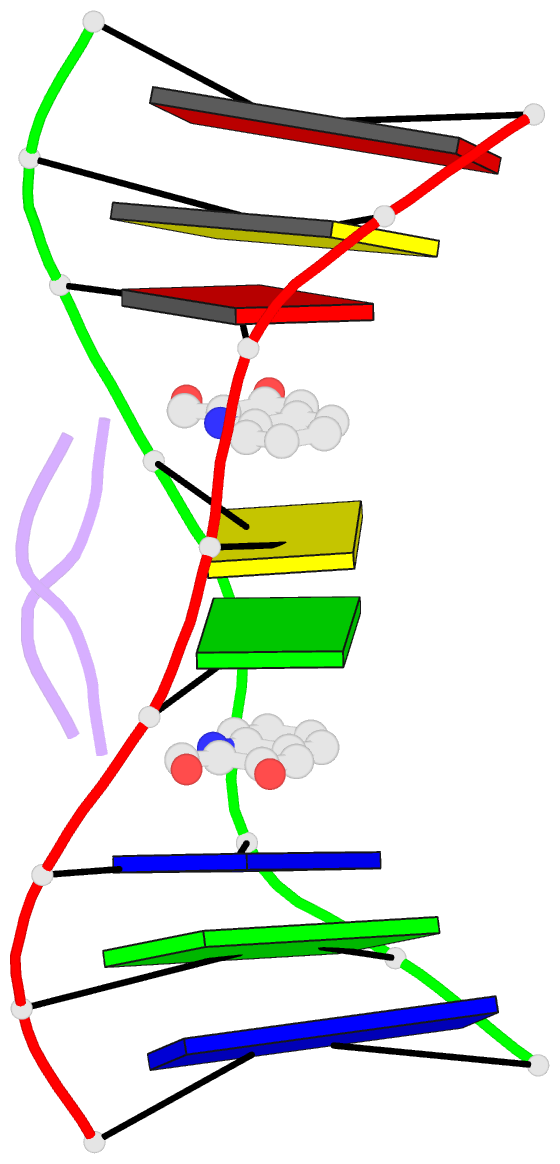
- Solution structure of a quinomycin bisintercalator-DNA complex
- Reference
- Chen H, Patel DJ (1995): "Solution Structure of a Quinomycin Bisintercalator-DNA Complex." J.Mol.Biol., 246, 164. doi: 10.1006/JMBI.1994.0074.
- Abstract
- The quinomycin antibiotic UK-63052 (designated QN) exhibits a chemical structure related to the antibiotic echinomycin which is known to bisintercalate into DNA. Common features among these antibiotics include two heterocyclic aromatic ring systems propagating from a cross-bridged cyclic octadepsipeptide scaffold. We report on the solution structure of the QN-d(A1-C2-A3-C4-G5-T6-G7-T8) complex (one QN molecule per duplex) based on a combined NMR-molecular dynamics study including intensity-based refinement. The 3-hydroxy quinaldic acid rings bisintercalate into the duplex at (A3-C4).(G5-T6) steps and stack with flanking Watson-Crick A3.T6 and C4.G5 base-pairs. The intercalation sites at (A3-C4).(G5-T6) steps are wedge-shaped and unwound, with significant unwinding also observed at the (C4-C5).(C4-G5) step bracketed between the intercalation sites. The cross-bridged cyclic octadepsipeptide is positioned in the minor groove with the methyl groups on its Ala and NMe-MCp residues directed towards and making van der Waals contacts with the minor groove edge of the duplex. A pair of adjacent intermolecular hydrogen bonds between the Ala backbone atoms and the G5 minor groove edge (Ala-NH to G5-N(3) and G5-NH2e to Ala-CO) account for the sequence specificity associated with complex formation. The solution structure of the QN-DNA oligomer complex, which contains only Watson-Crick base-pairs flanking the bisintercalation site, is compared with the crystal structure of the related echinomycin-DNA oligomer complex, which contains Hoogsteen base-pairs on either side of the bisintercalation site.