Summary information and primary citation
- PDB-id
- 1a4t; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-RNA
- Method
- NMR
- Summary
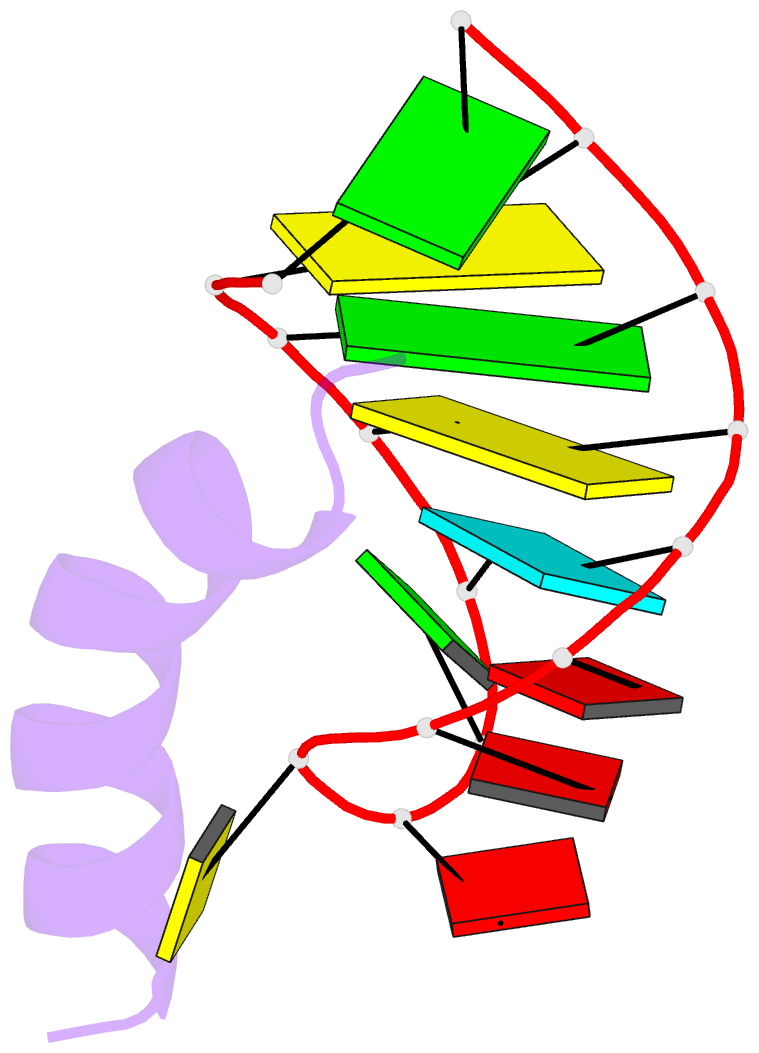
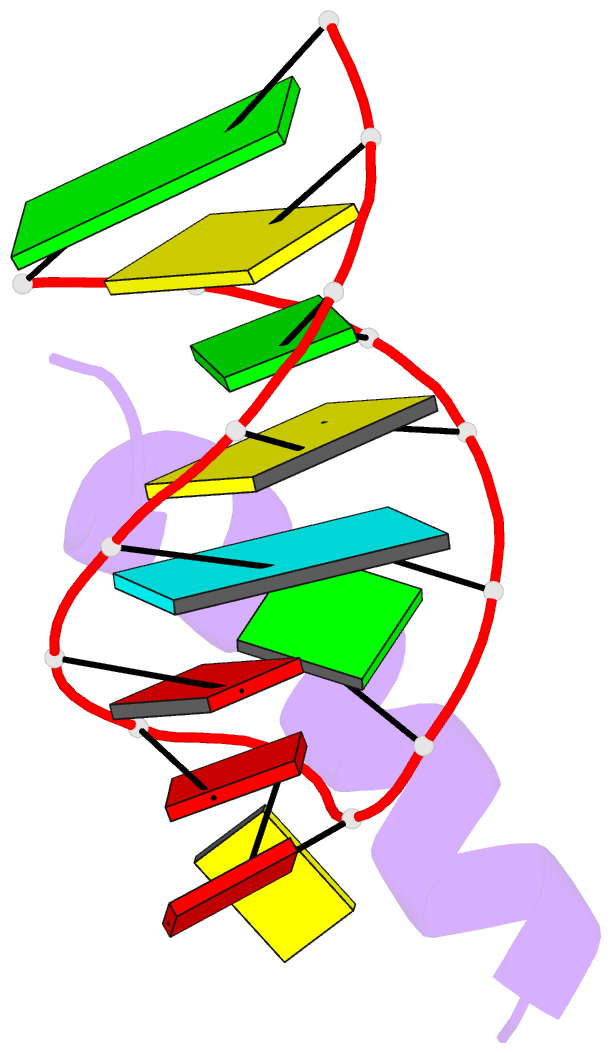
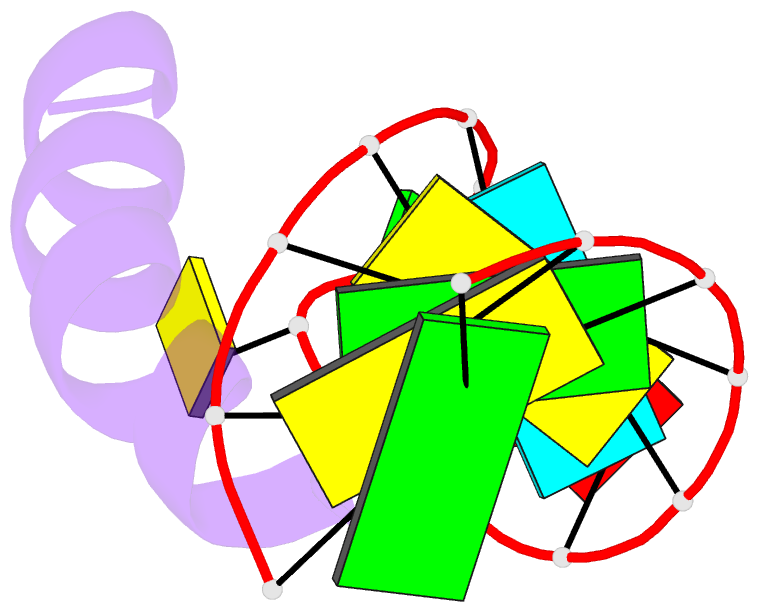
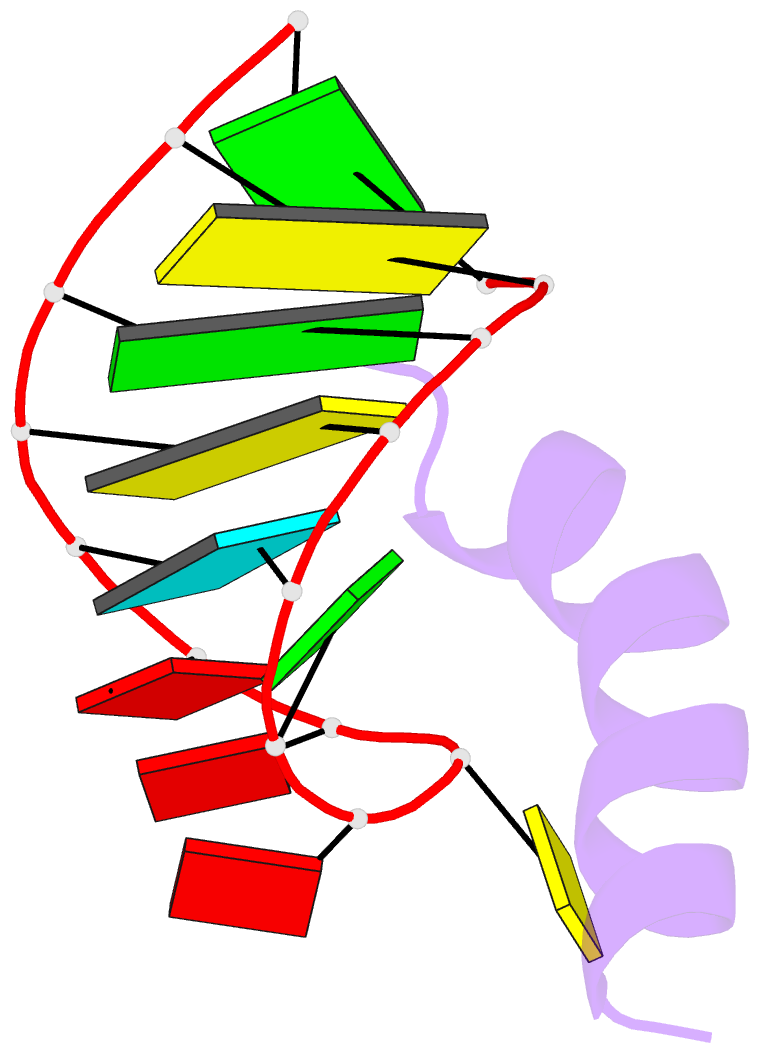
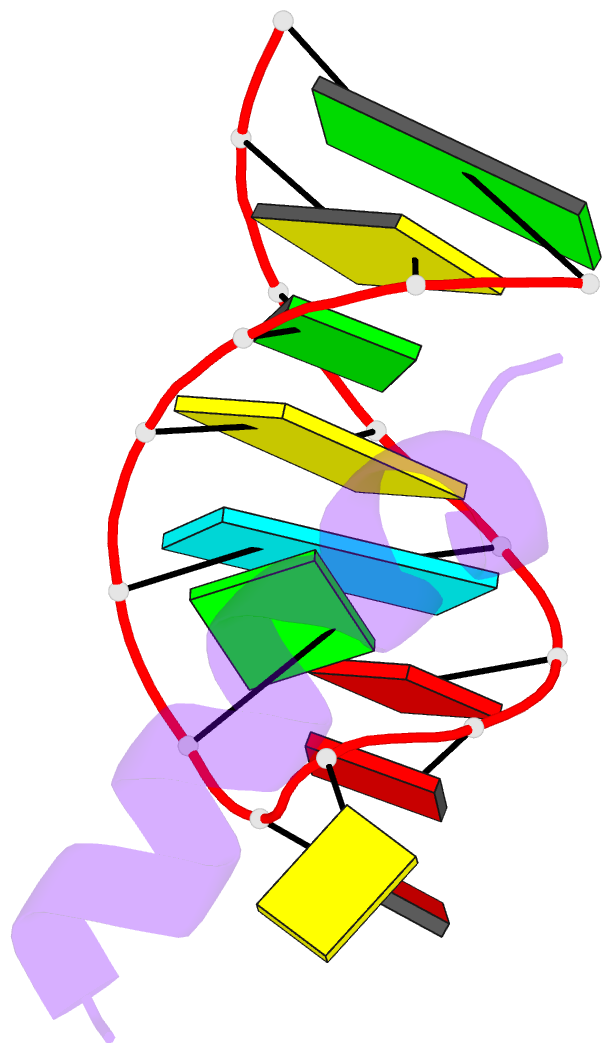
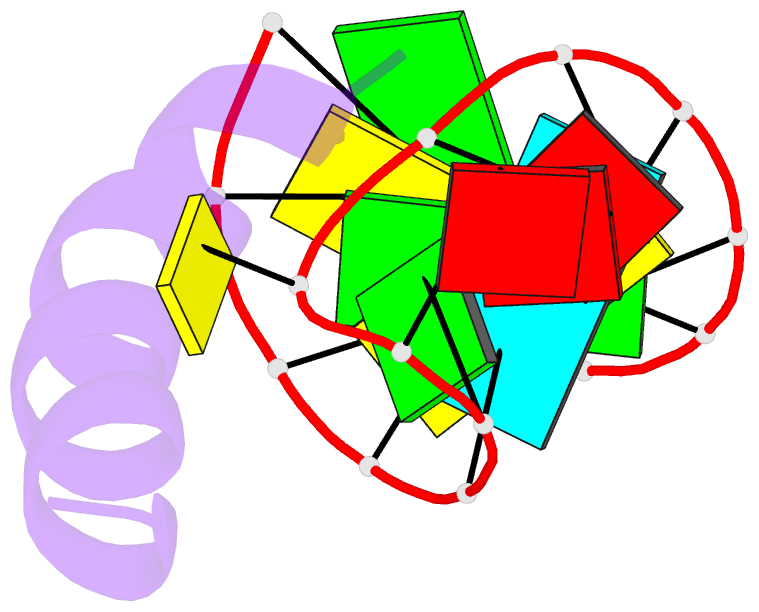
- Solution structure of phage p22 n peptide-box b RNA complex, NMR, 20 structures
- Reference
- Cai Z, Gorin A, Frederick R, Ye X, Hu W, Majumdar A, Kettani A, Patel DJ (1998): "Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex." Nat.Struct.Biol., 5, 203-212. doi: 10.1038/nsb0398-203.
- Abstract
- We have determined the solution structure of a 15-mer boxB RNA hairpin complexed with a 20-mer basic peptide of the N protein involved in bacteriophage P22 transcriptional antitermination. Complex formation involves adaptive binding with the N peptide adopting a bent alpha-helical conformation that packs tightly through hydrophobic and electrostatic interactions against the major groove face of the boxB RNA hairpin, orienting the open opposite face for potential interactions with host factors and/or RNA polymerase. Four nucleotides in the boxB RNA hairpin pentaloop form a stable GNRA like tetraloop structural scaffold on complex formation, allowing the looped out fifth nucleotide to make extensive hydrophobic contacts with the bound peptide. The guanidinium group of a key arginine is hydrogen-bonded to the guanine in a loop-closing sheared G.A mismatch and to adjacent backbone phosphates. The identified intermolecular contacts account for the consequences of N peptide and boxB RNA mutations on bacteriophage transcriptional antitermination.