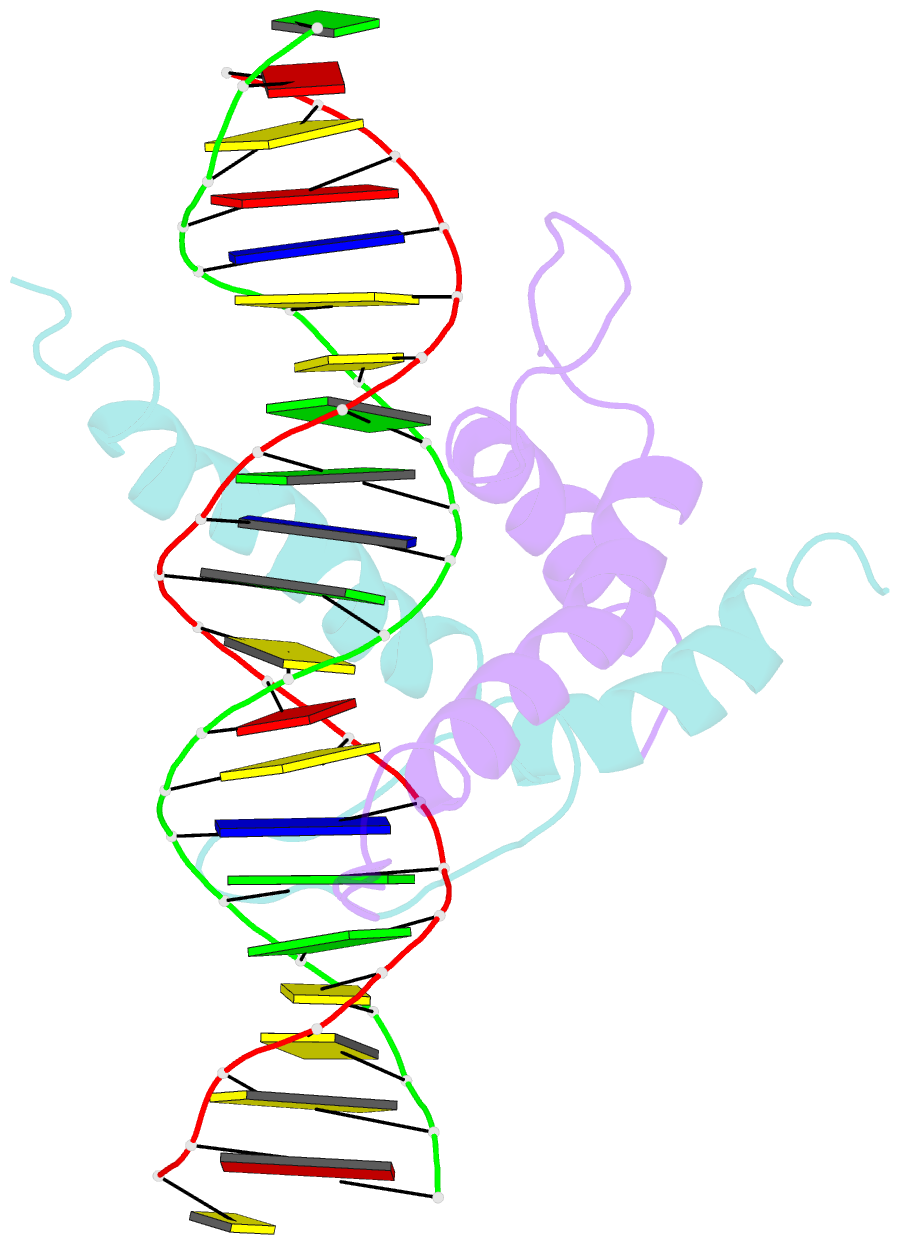
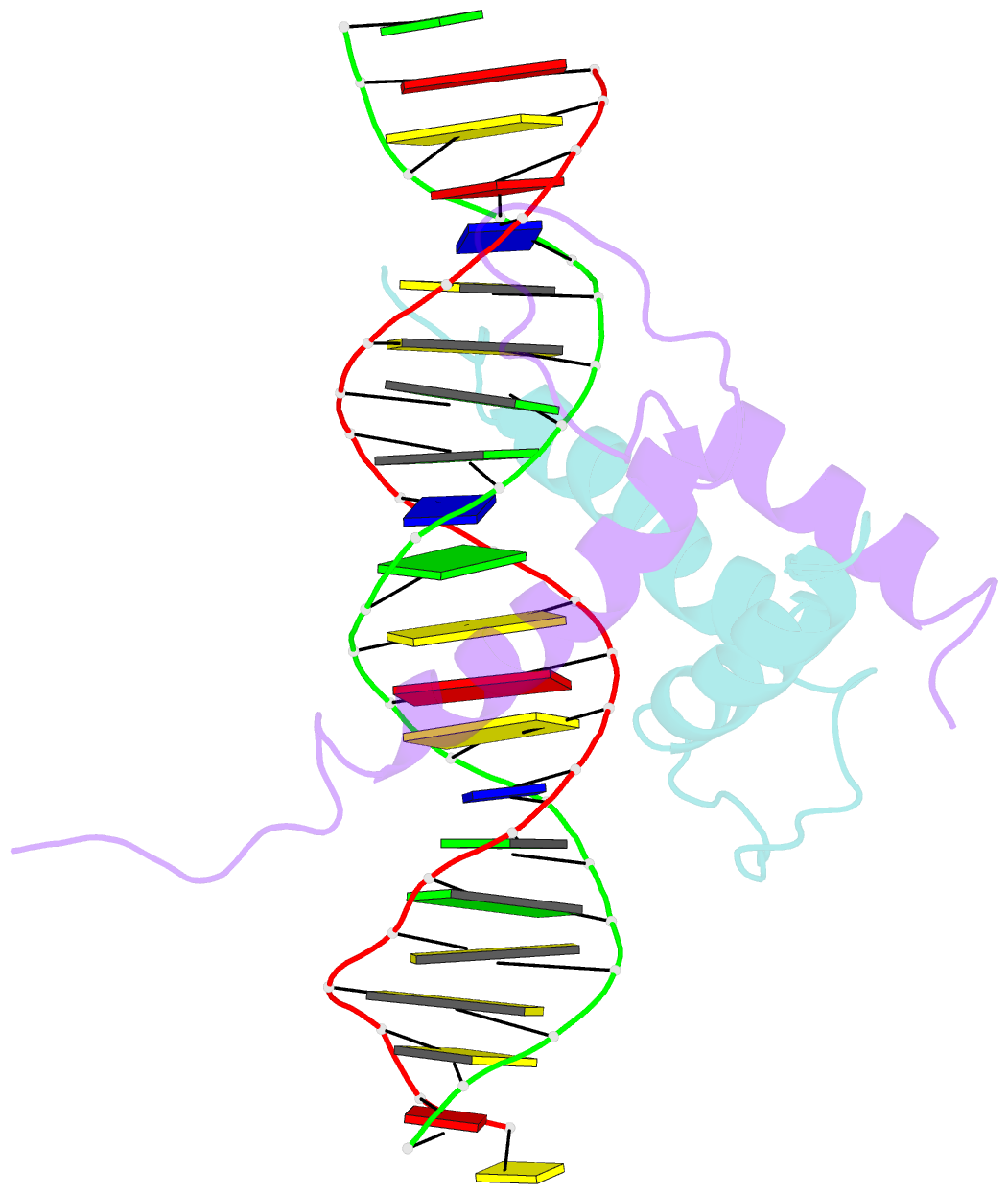
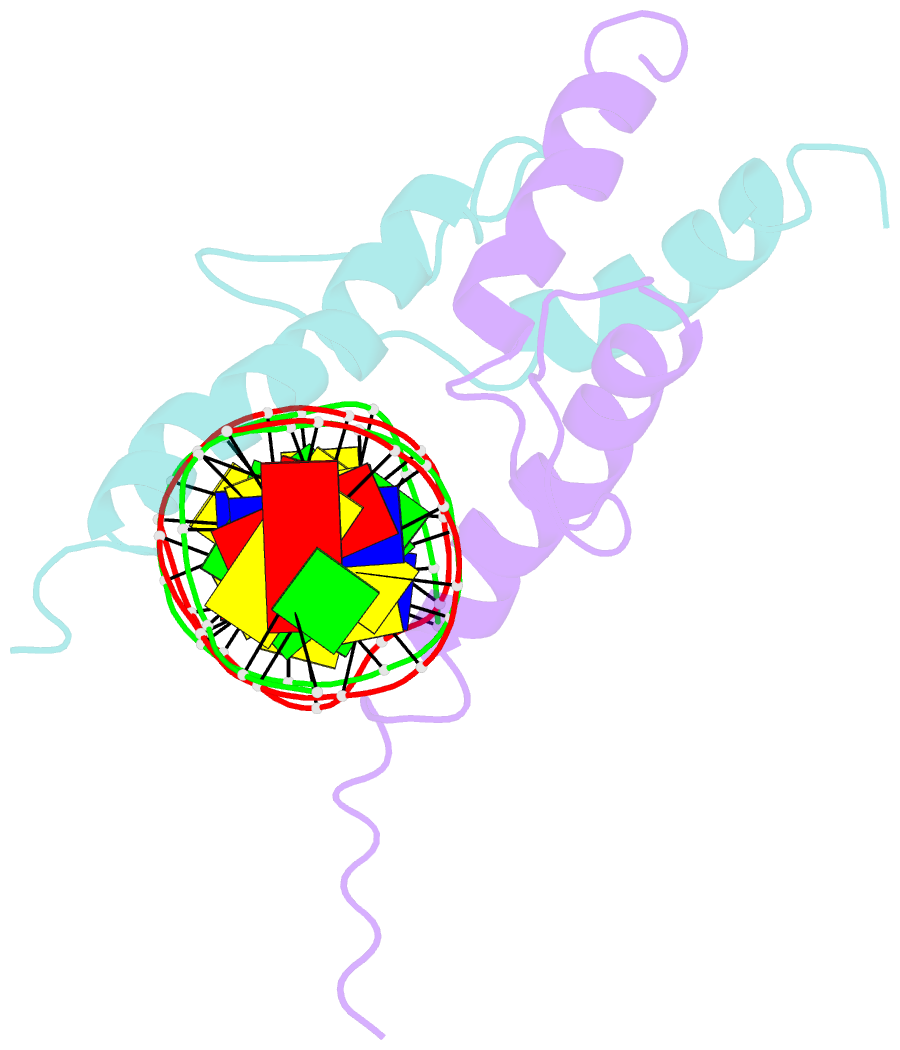
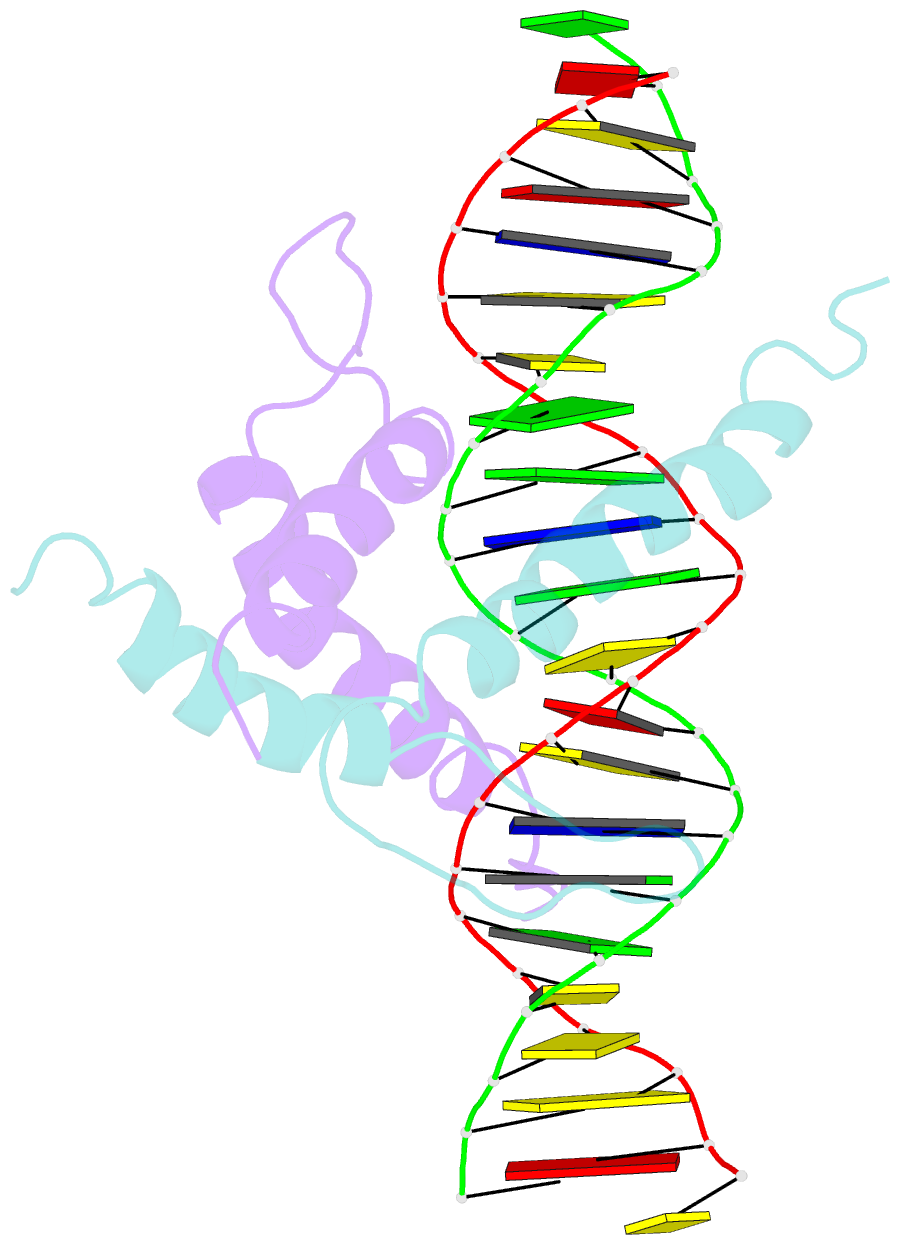
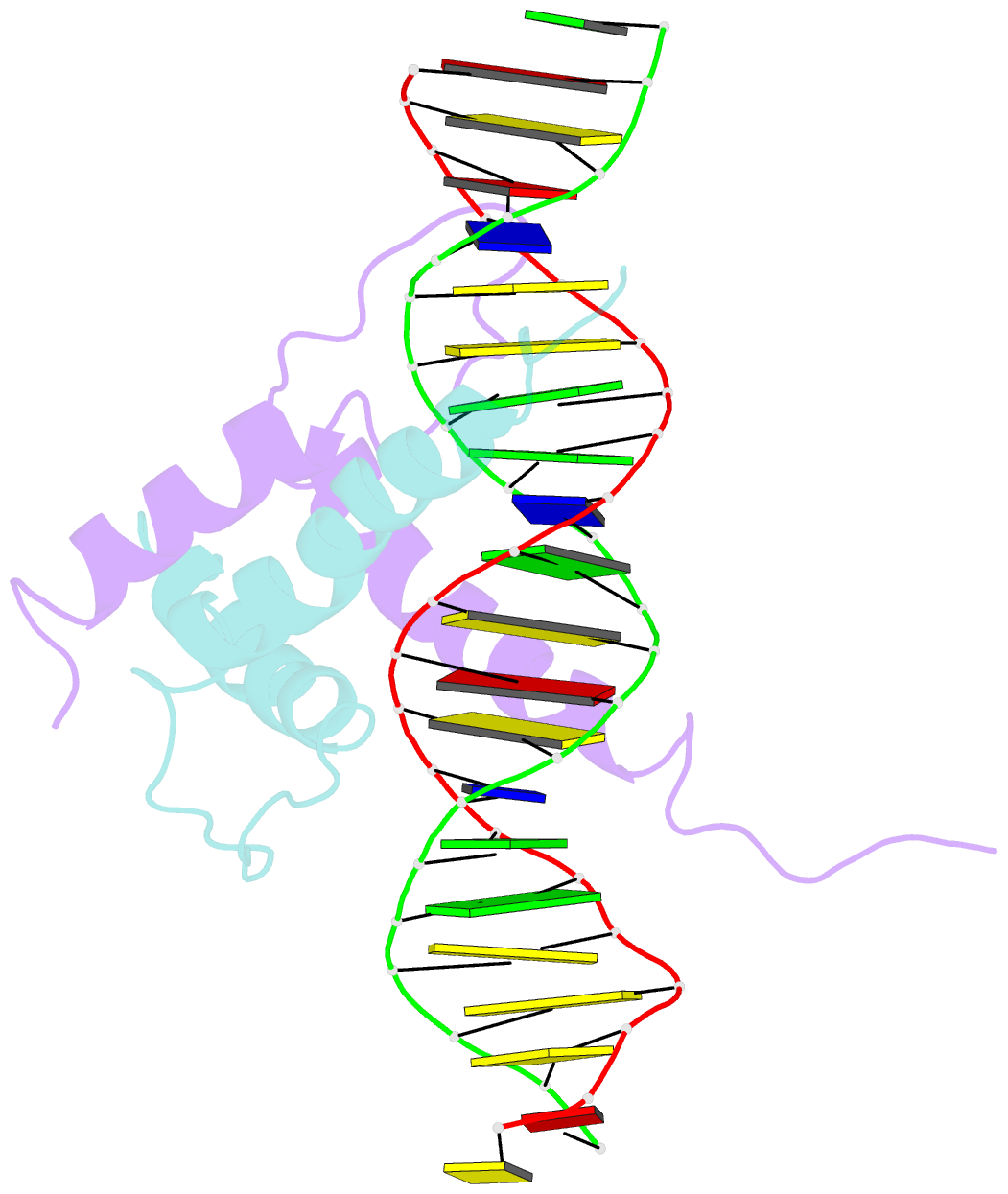
Summary information and primary citation
- PDB-id
- 1an4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.9 Å)
- Summary
- Structure and function of the b-hlh-z domain of usf
- Reference
- Ferre-D'Amare AR, Pognonec P, Roeder RG, Burley SK (1994): "Structure and function of the b/HLH/Z domain of USF." EMBO J., 13, 180-189.
- Abstract
- The basic/helix-loop-helix/leucine zipper (b/HLH/Z) transcription factor upstream stimulatory factor (USF) and its isolated DNA binding domain undergo a random coil to alpha-helix folding transition on recognizing their cognate DNA. The USF b/HLH cocrystal structure resembles the structure of the b/HLH/Z domain of the homologous protein Max and reveals (i) that the truncated, b/HLH DNA binding domain homodimerizes, forming a parallel, left-handed four-helix bundle, and (ii) that the basic region becomes alpha-helical on binding to the major groove of the DNA sequence CACGTG. Hydrodynamic measurements show that the b/HLH/Z DNA binding domain of USF exists as a bivalent homotetramer. This tetramer forms at the USF physiological intranuclear concentration, and depends on the integrity of the leucine zipper motif. The ability to bind simultaneously to two independent sites suggests a role in DNA looping for the b/HLH/Z and Myc-related families of eukaryotic transcription factors.