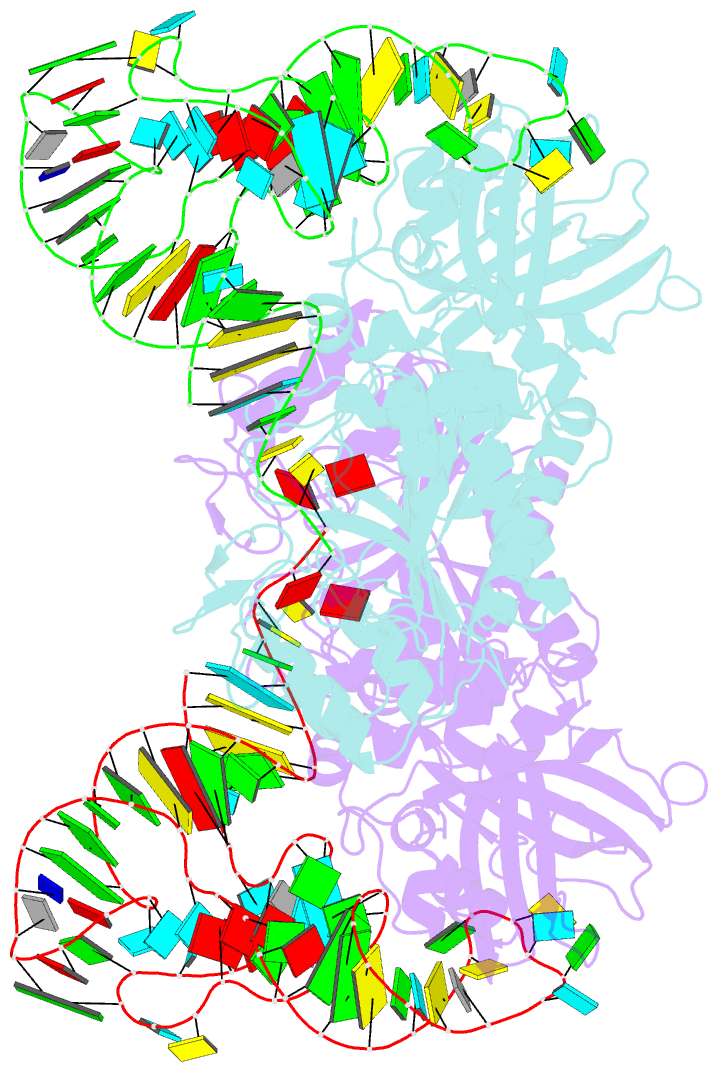
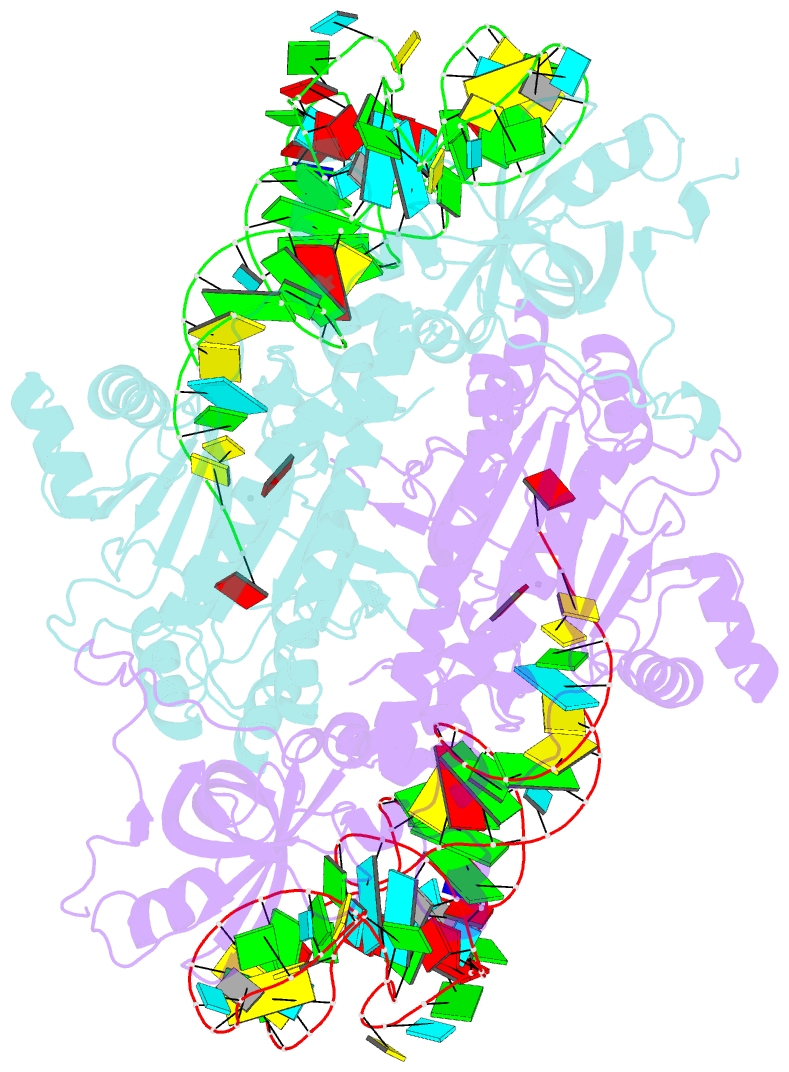
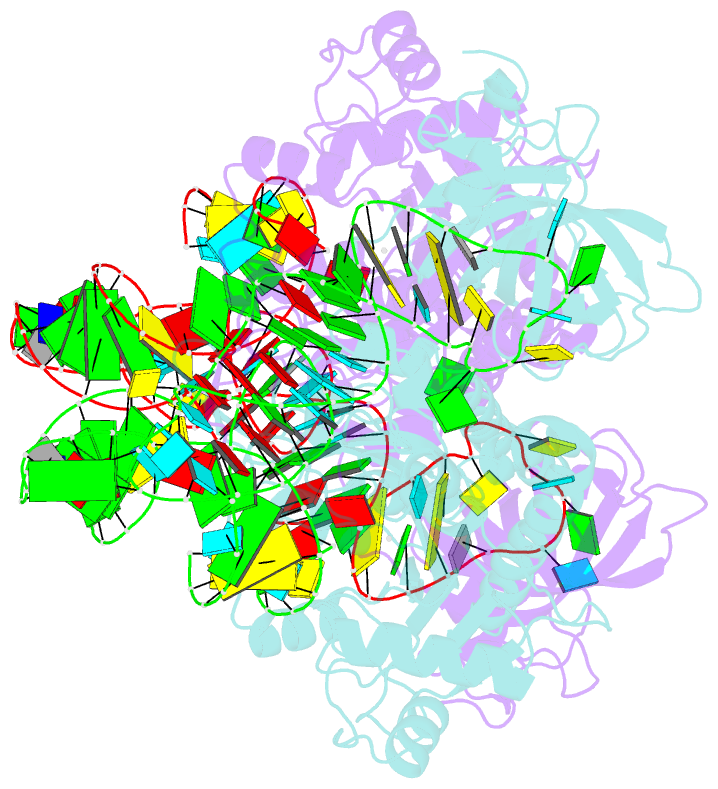
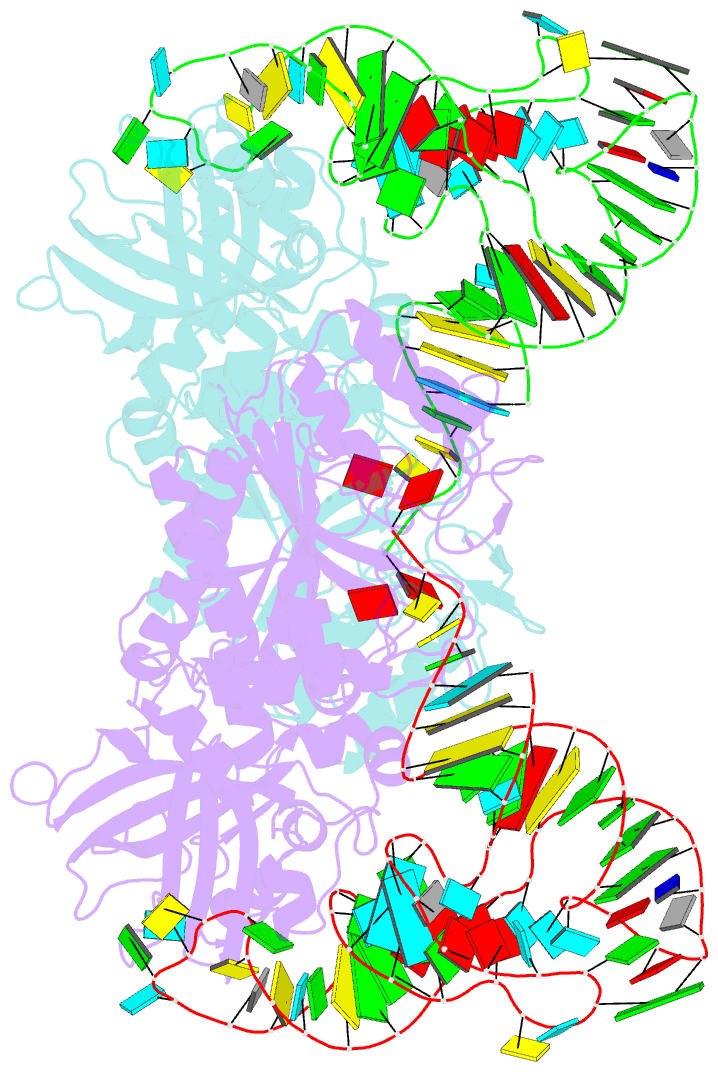
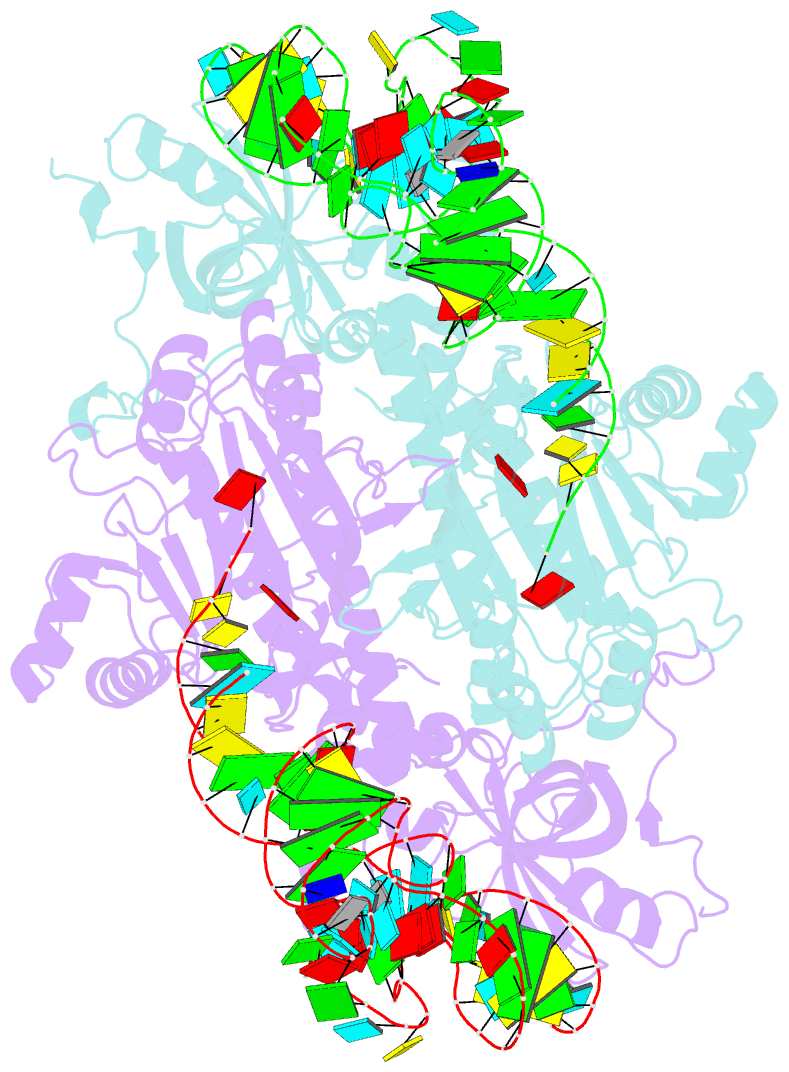
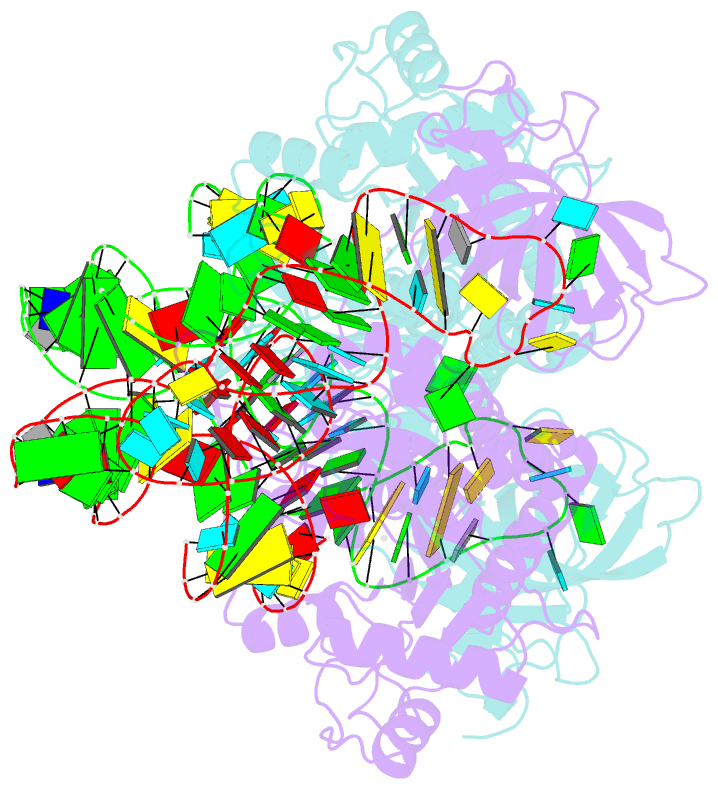
Summary information and primary citation
- PDB-id
- 1asz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- complex (aminoacyl-trna synthase-trna)
- Method
- X-ray (3.0 Å)
- Summary
- The active site of yeast aspartyl-trna synthetase: structural and functional aspects of the aminoacylation reaction
- Reference
- Cavarelli J, Eriani G, Rees B, Ruff M, Boeglin M, Mitschler A, Martin F, Gangloff J, Thierry JC, Moras D (1994): "The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction." EMBO J., 13, 327-337.
- Abstract
- The crystal structures of the various complexes formed by yeast aspartyl-tRNA synthetase (AspRS) and its substrates provide snapshots of the active site corresponding to different steps of the aminoacylation reaction. Native crystals of the binary complex tRNA-AspRS were soaked in solutions containing the two other substrates, ATP (or its analog AMPPcP) and aspartic acid. When all substrates are present in the crystal, this leads to the formation of the aspartyl-adenylate and/or the aspartyl-tRNA. A class II-specific pathway for the aminoacylation reaction is proposed which explains the known functional differences between the two classes while preserving a common framework. Extended signature sequences characteristic of class II aaRS (motifs 2 and 3) constitute the basic functional unit. The ATP molecule adopts a bent conformation, stabilized by the invariant Arg531 of motif 3 and a magnesium ion coordinated to the pyrophosphate group and to two class-invariant acidic residues. The aspartic acid substrate is positioned by a class II invariant acidic residue, Asp342, interacting with the amino group and by amino acids conserved in the aspartyl synthetase family. The amino acids in contact with the substrates have been probed by site-directed mutagenesis for their functional implication.