Summary information and primary citation
- PDB-id
- 1b23; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation-RNA
- Method
- X-ray (2.6 Å)
- Summary
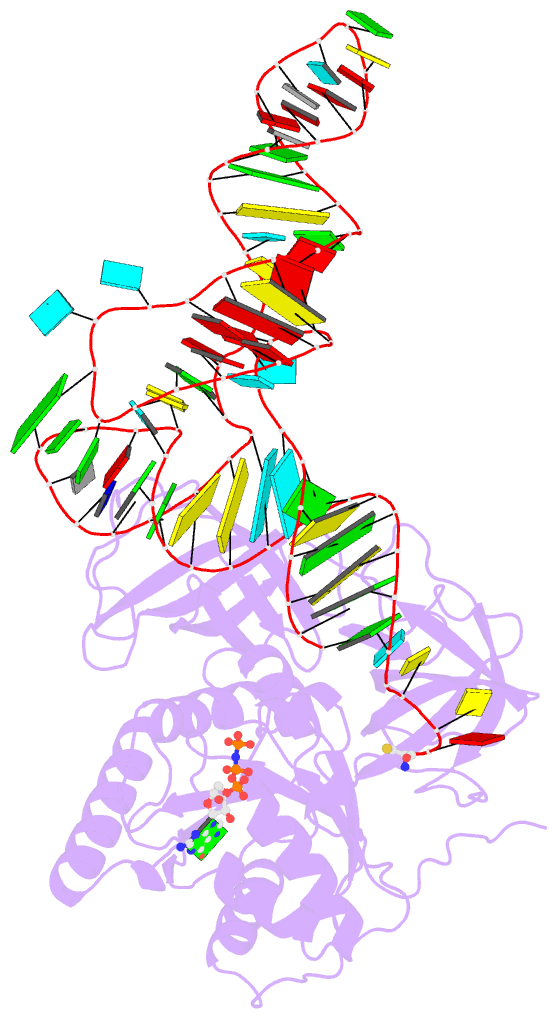
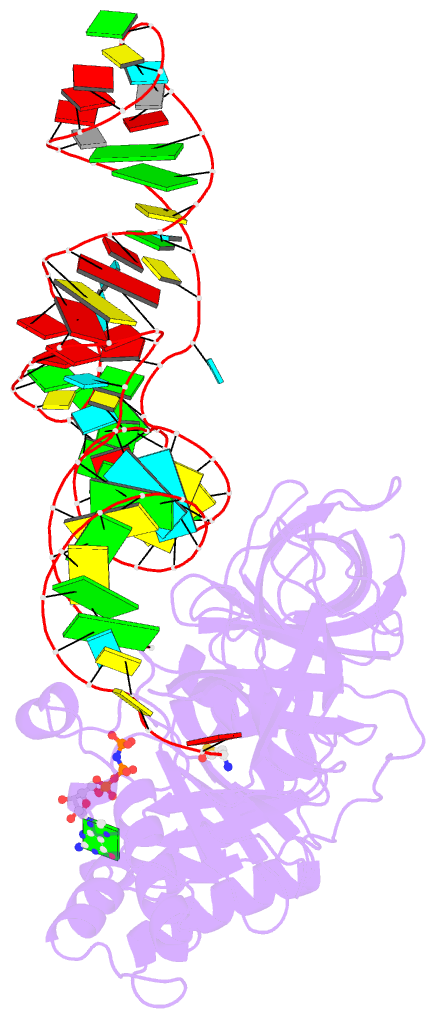
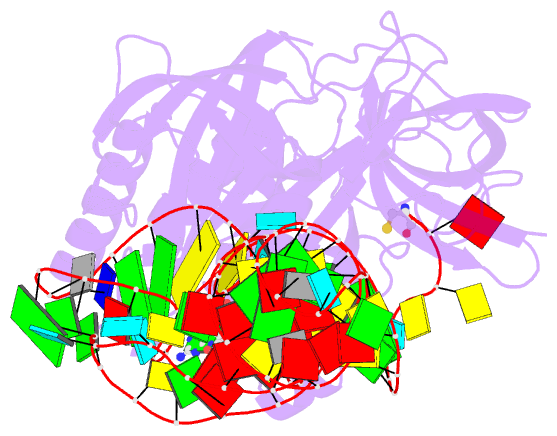
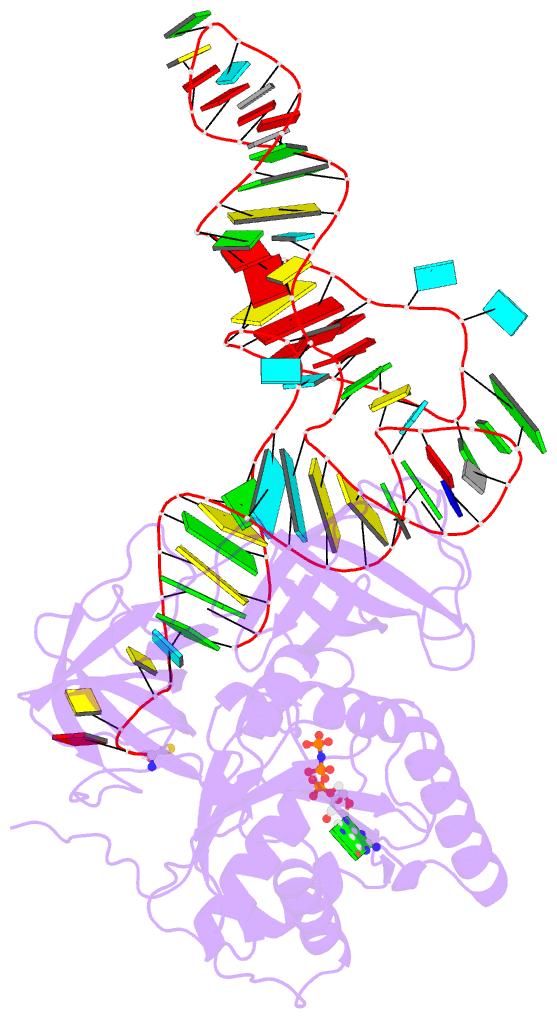
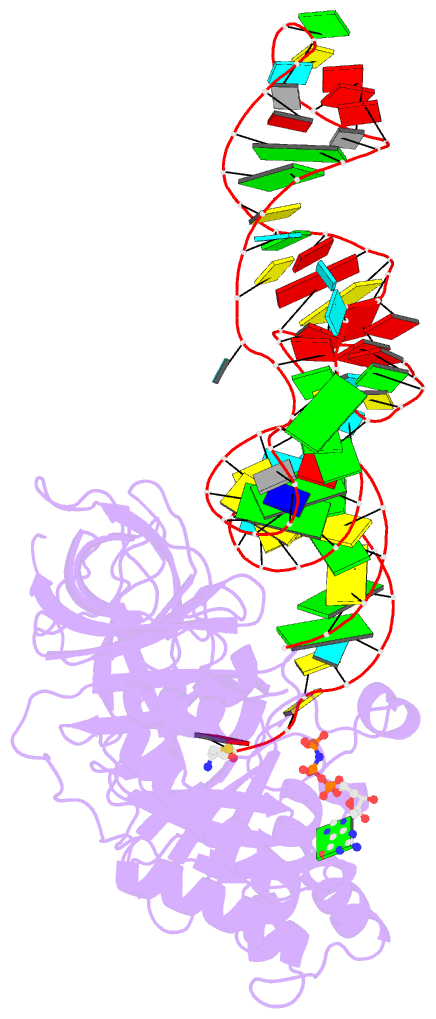
- E. coli cysteinyl-trna and t. aquaticus elongation factor ef-tu:gtp ternary complex
- Reference
- Nissen P, Thirup S, Kjeldgaard M, Nyborg J (1999): "The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA." Structure Fold.Des., 7, 143-156. doi: 10.1016/S0969-2126(99)80021-5.
- Abstract
- Background: . The translation elongation factor EF-Tu in its GTP-bound state forms a ternary complex with any aminoacylated tRNA (aa-tRNA), except initiator tRNA and selenocysteinyl-tRNA. This complex delivers aa-tRNA to the ribosomal A site during the elongation cycle of translation. The crystal structure of the yeast Phe-tRNAPhe ternary complex with Thermus aquaticus EF-Tu-GDPNP (Phe-TC) has previously been determined as one representative of this general yet highly discriminating complex formation.
Results: The ternary complex of Escherichia coli Cys-tRNACys and T. aquaticus EF-Tu-GDPNP (Cys-TC) has been solved and refined at 2.6 degrees resolution. Conserved and variable features of the aa-tRNA recognition and binding by EF-Tu-GTP have been revealed by comparison with the Phe-TC structure. New tertiary interactions are observed in the tRNACys structure. A 'kissing complex' is observed in the very close crystal packing arrangement.
Conclusions: The recognition of Cys-tRNACys by EF-Tu-GDPNP is restricted to the aa-tRNA motif previously identified in Phe-TC and consists of the aminoacylated 3' end, the phosphorylated 5' end and one side of the acceptor stem and T stem. The aminoacyl bond is recognized somewhat differently, yet by the same primary motif in EF-Tu, which suggests that EF-Tu adapts to subtle variations in this moiety among all aa-tRNAs. New tertiary interactions revealed by the Cys-tRNACys structure, such as a protonated C16:C59 pyrimidine pair, a G15:G48 'Levitt pair' and an s4U8:A14:A46 base triple add to the generic understanding of tRNA structure from sequence. The structure of the 'kissing complex' shows a quasicontinuous helix with a distinct shape determined by the number of base pairs.