Summary information and primary citation
- PDB-id
- 1bc8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (1.93 Å)
- Summary
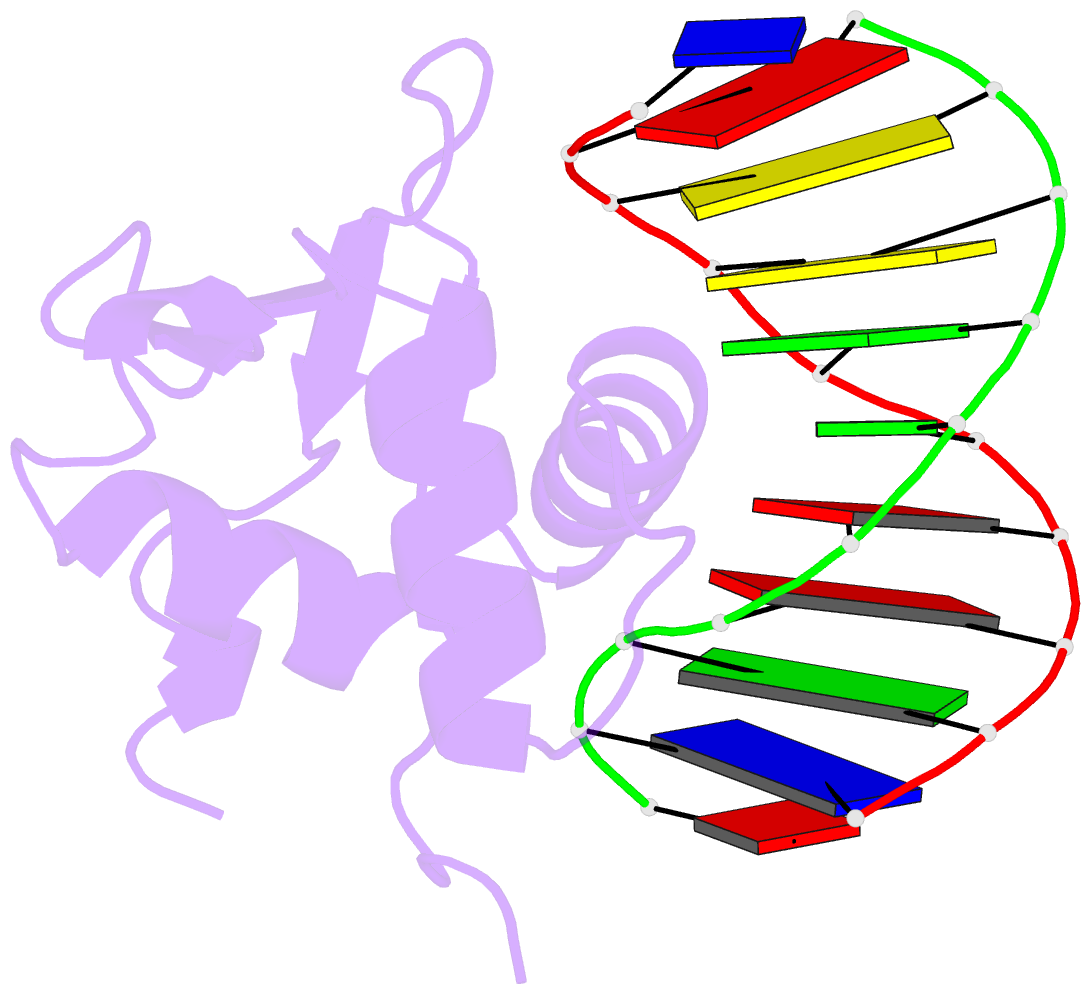
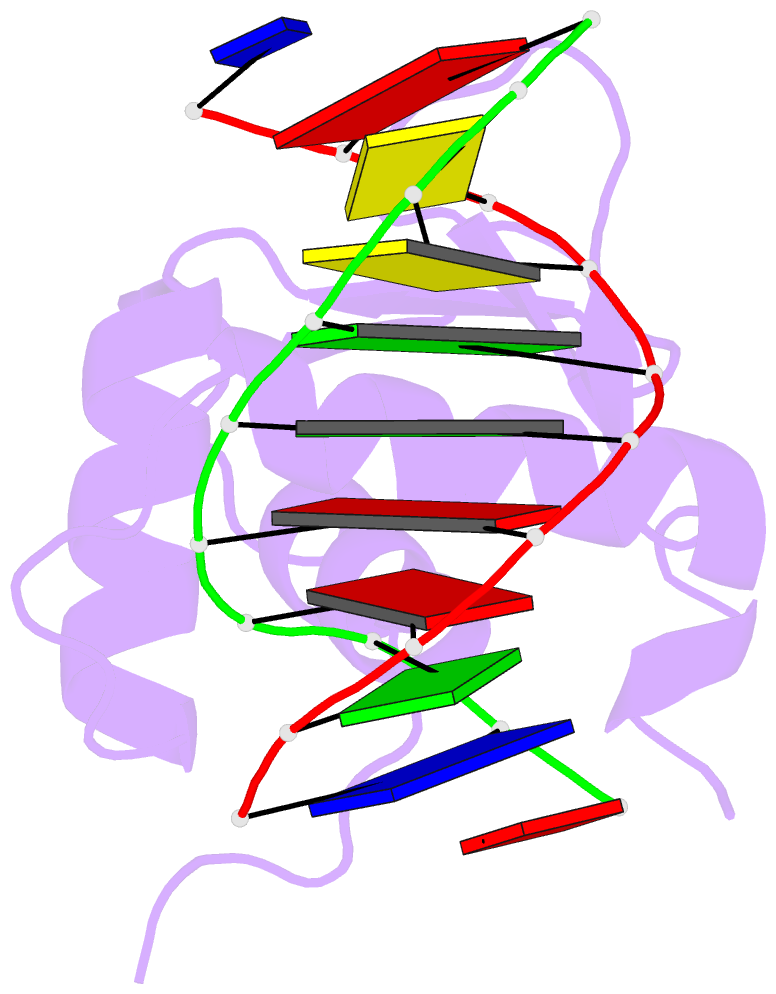
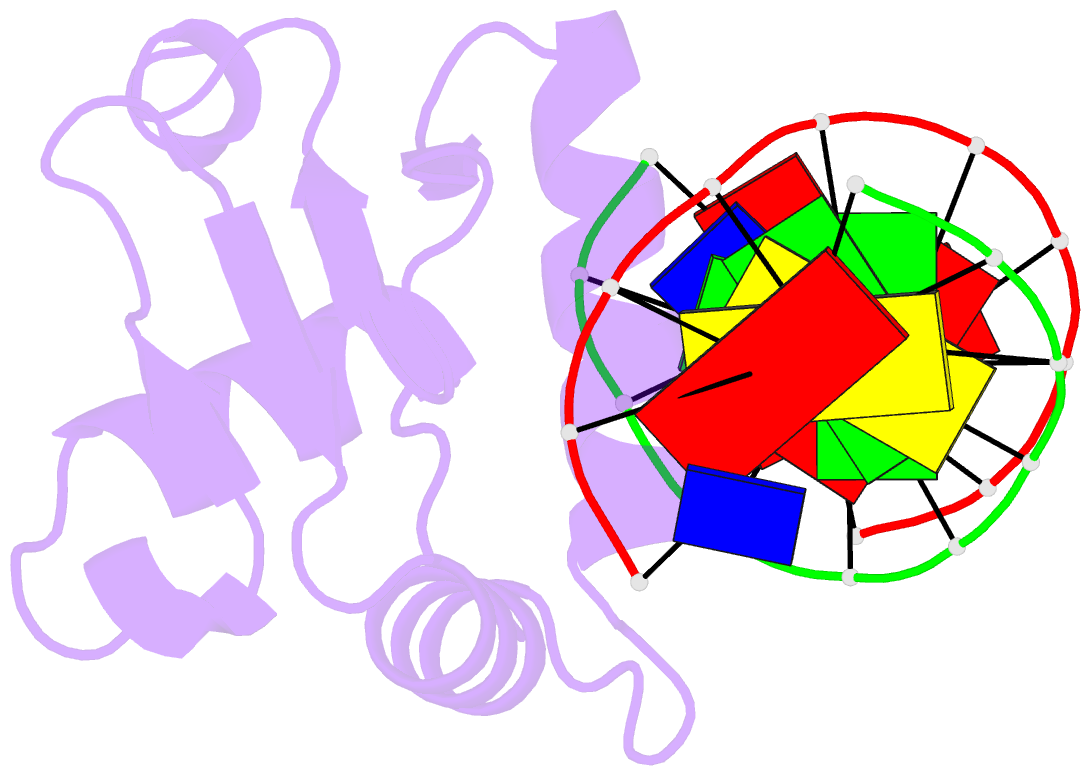
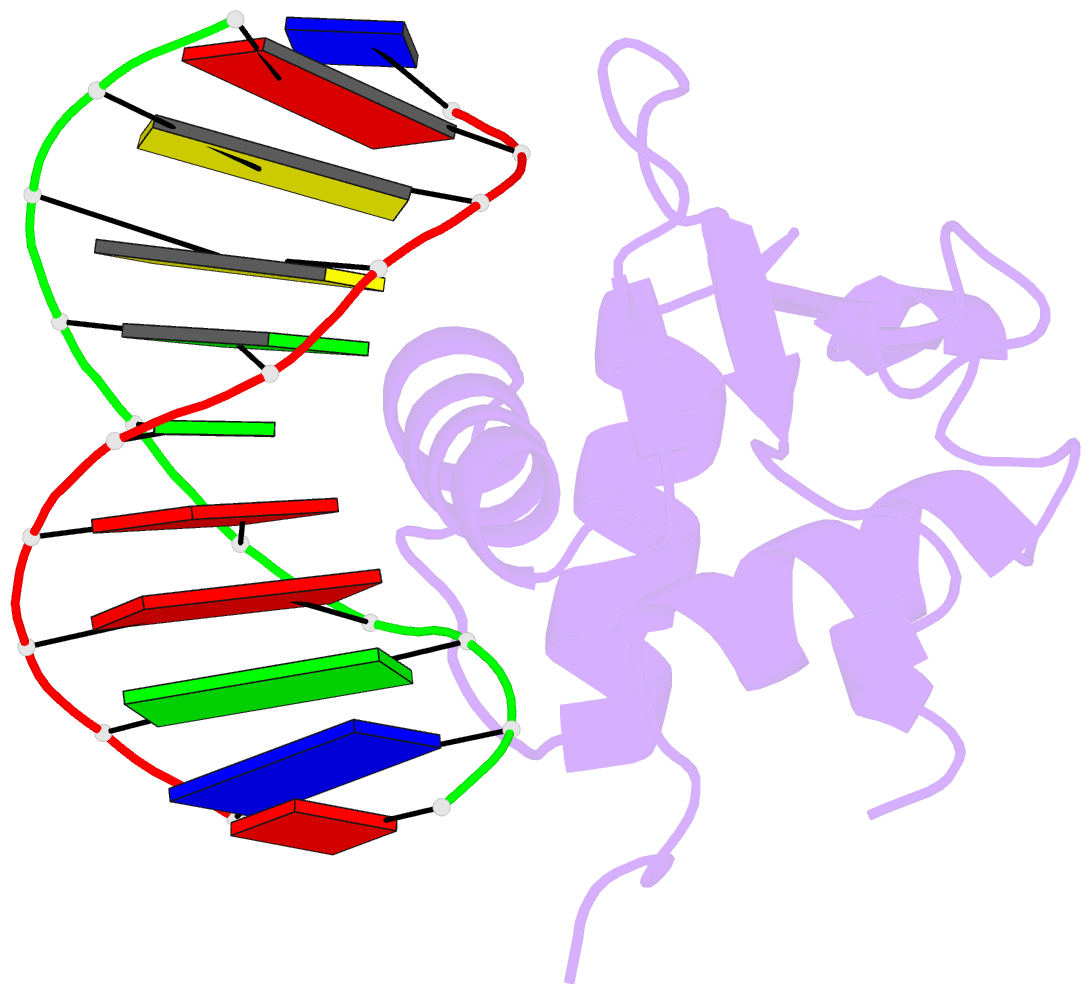
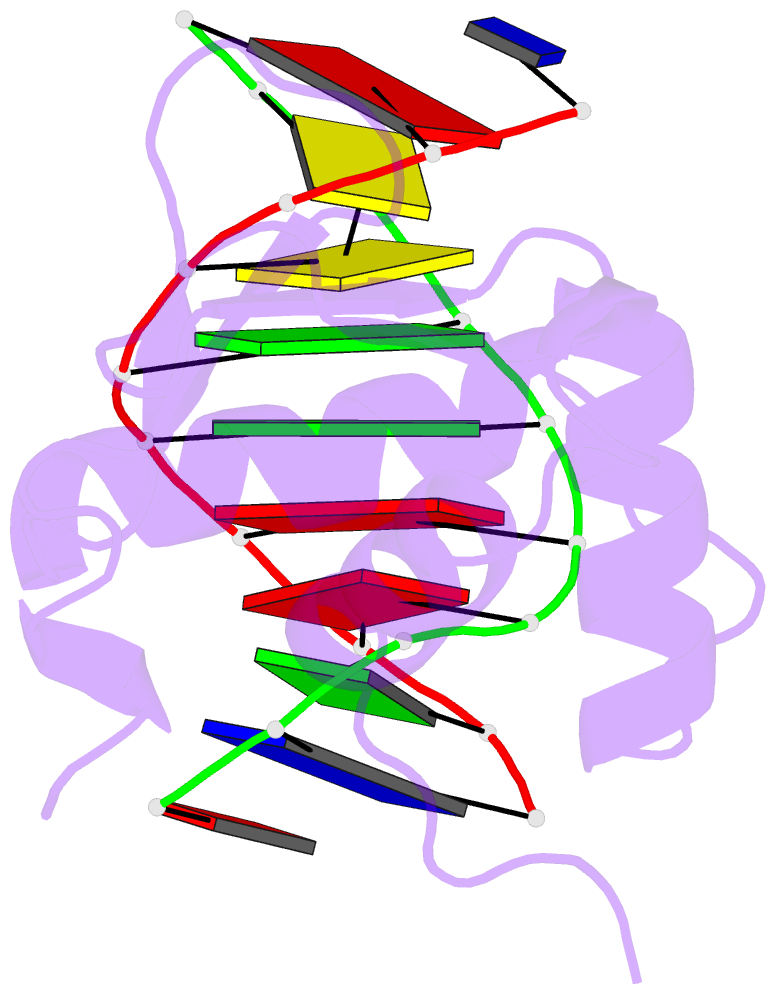
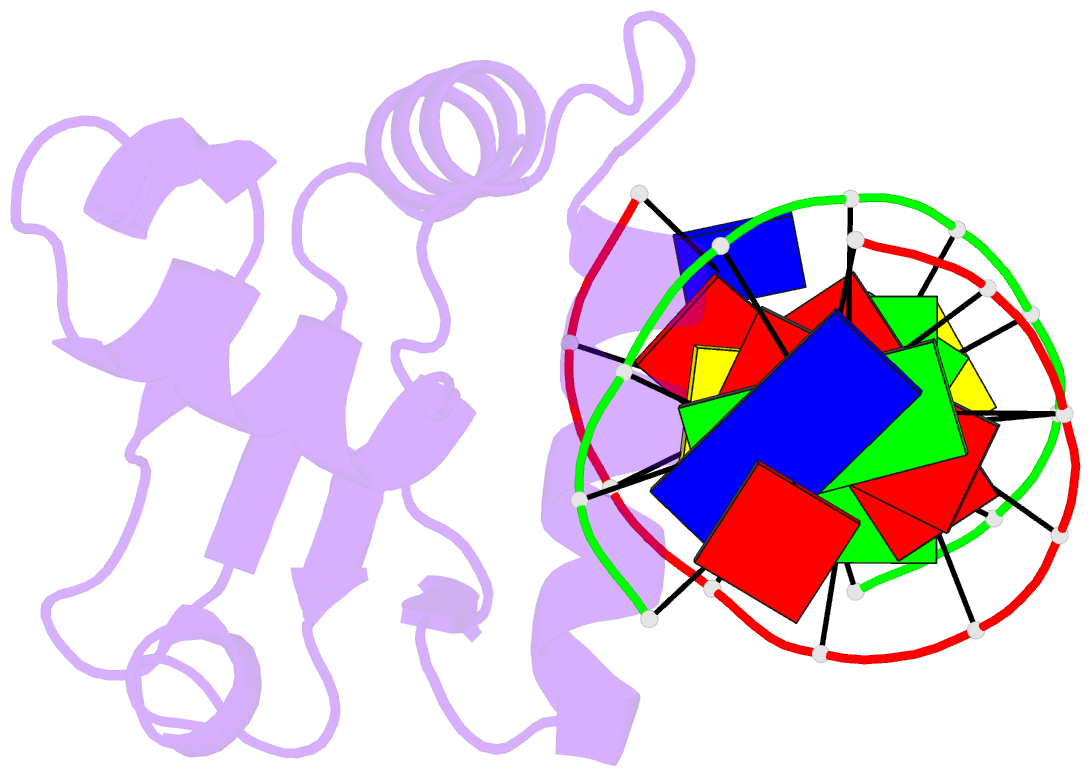
- Structures of sap-1 bound to DNA sequences from the e74 and c-fos promoters provide insights into how ets proteins discriminate between related DNA targets
- Reference
- Mo Y, Vaessen B, Johnston K, Marmorstein R (1998): "Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins." Mol.Cell, 2, 201-212. doi: 10.1016/S1097-2765(00)80130-6.
- Abstract
- SAP-1 is a member of the Ets transcription factors and cooperates with SRF protein to activate transcription of the c-fos protooncogene. The crystal structures of the conserved ETS domain of SAP-1 bound to DNA sequences from the E74 and c-fos promoters reveal that a set of conserved residues contact a GGA core DNA sequence. Discrimination for sequences outside this core is mediated by DNA contacts from conserved and nonconserved protein residues and sequence-dependent DNA structural properties characteristic of A-form DNA structure. Comparison with the related PU.1/DNA and GABPalpha/beta/DNA complexes provides general insights into DNA discrimination between Ets proteins. Modeling studies of a SAP-1/SRF/DNA complex suggest that SRF may modulate SAP-1 binding to DNA by interacting with its ETS domain.