Summary information and primary citation
- PDB-id
- 1bhm; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.2 Å)
- Summary
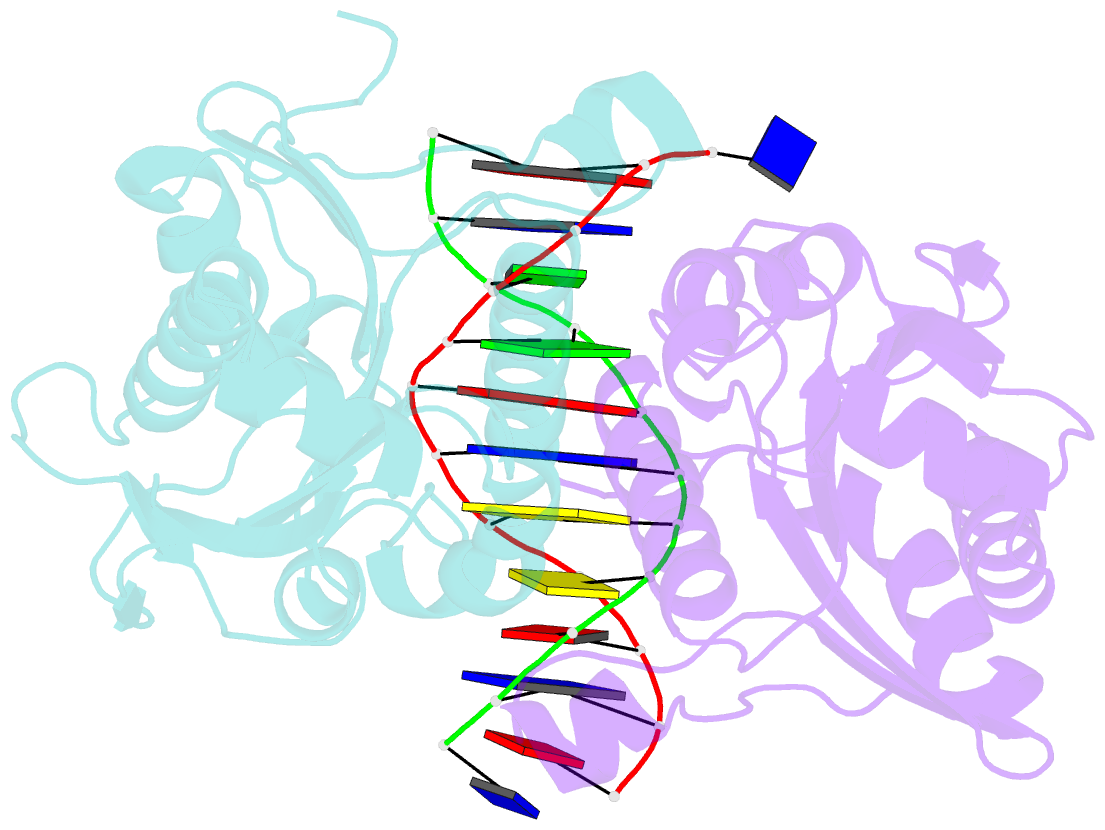
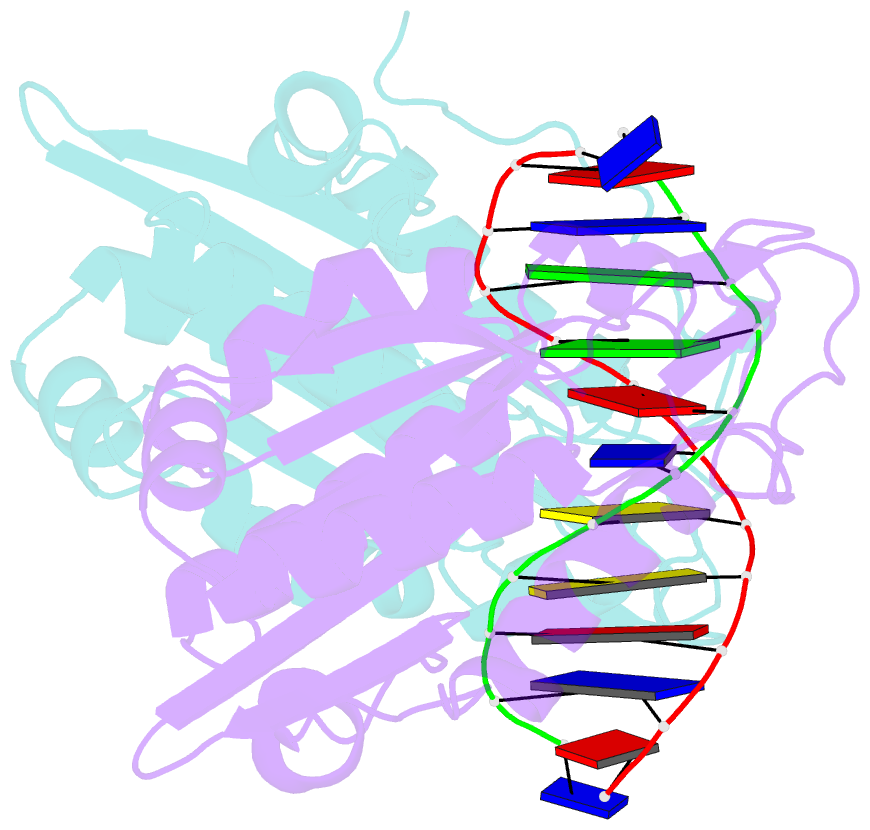
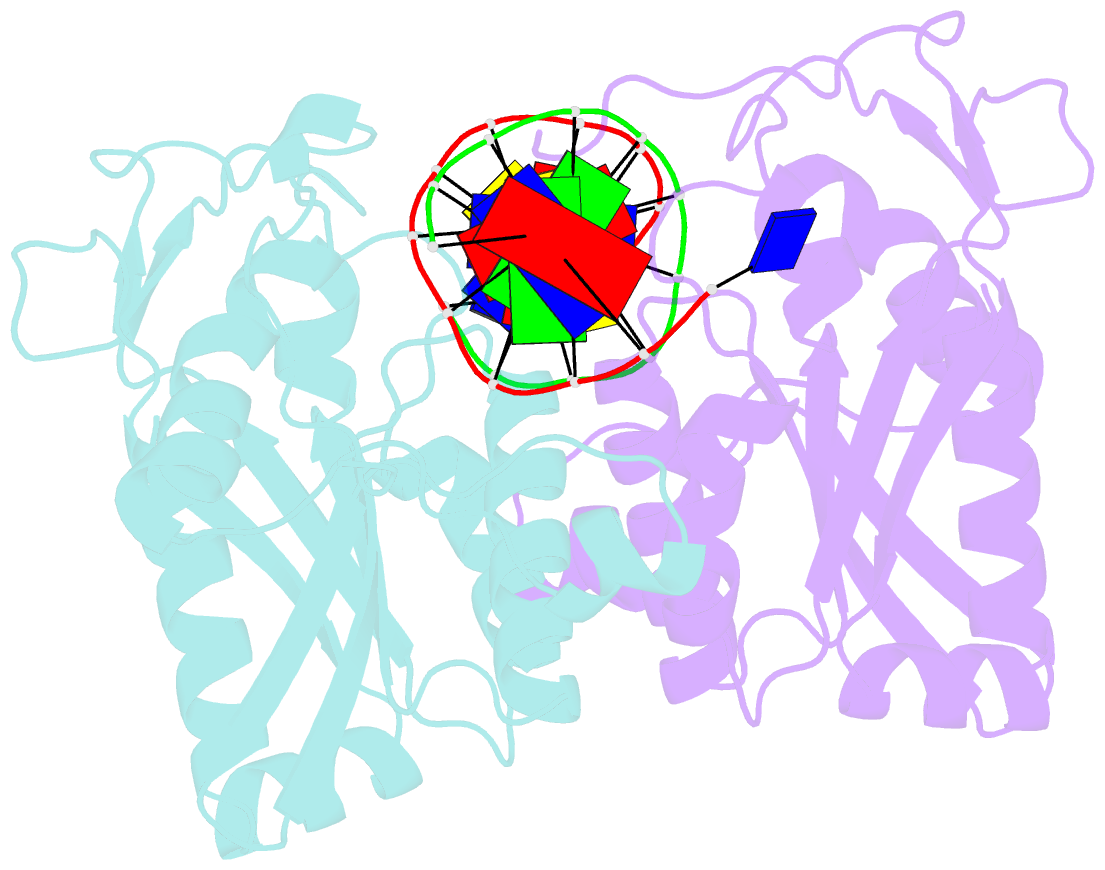
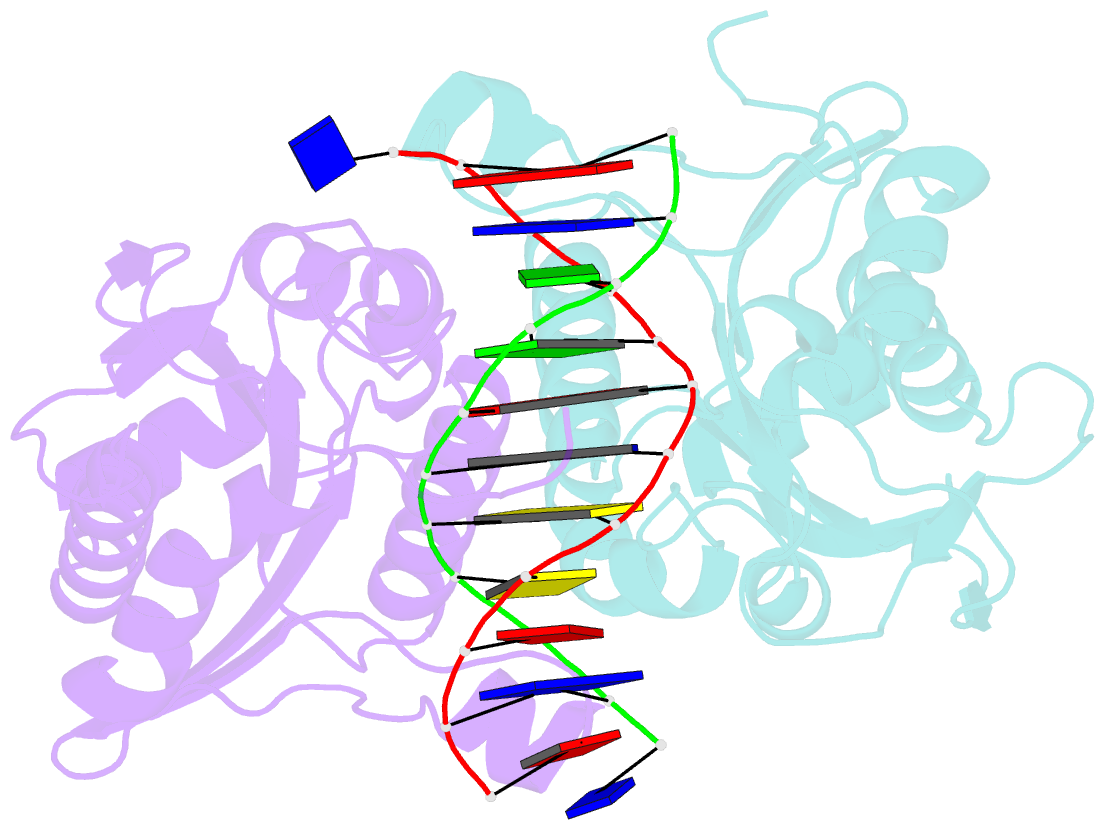
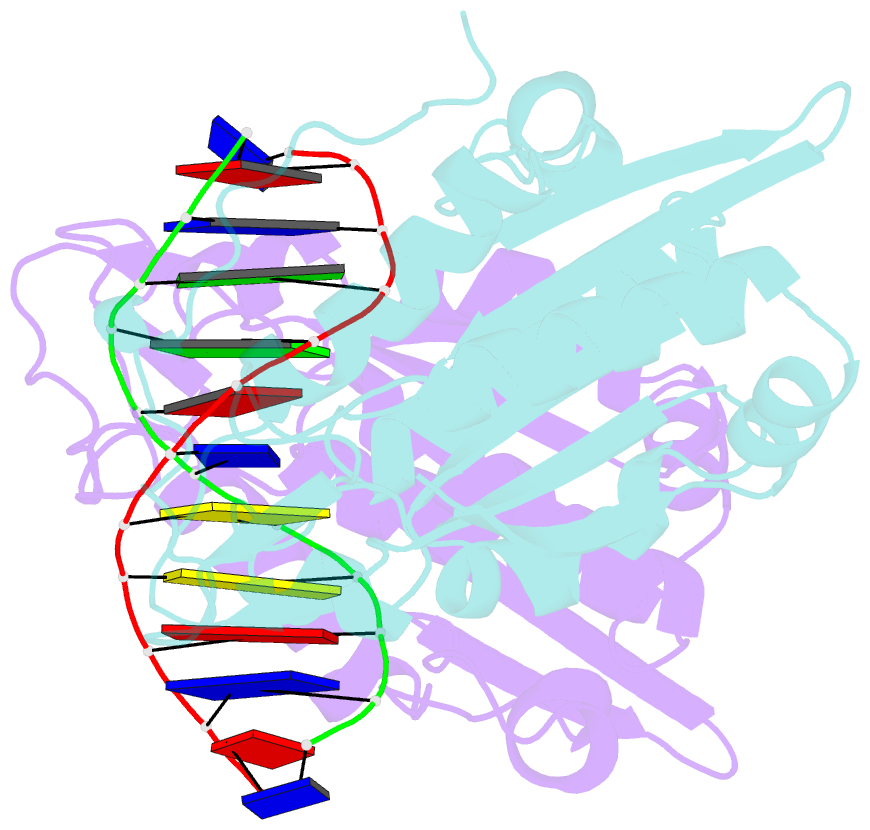
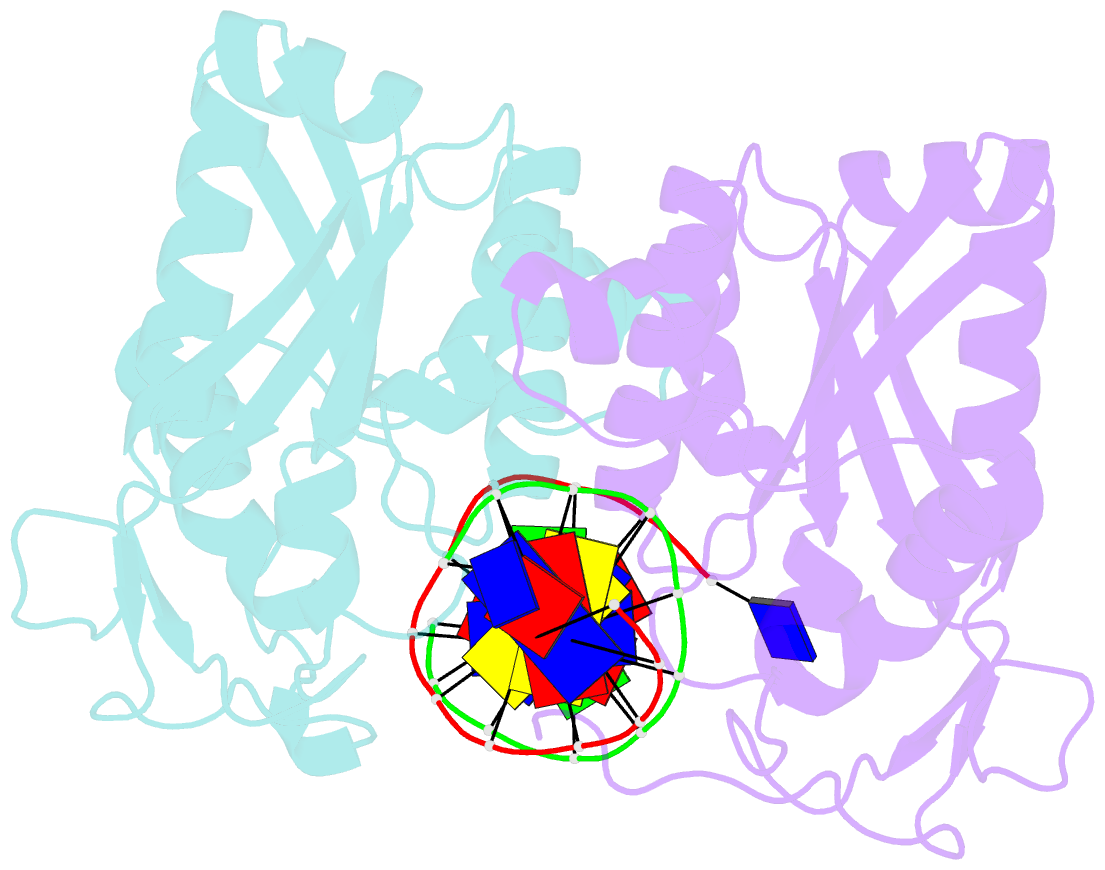
- Restriction endonuclease bamhi complex with DNA
- Reference
- Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK (1995): "Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding." Science, 269, 656-663.
- Abstract
- The crystal structure of restriction endonuclease Bam HI complexed to DNA has been determined at 2.2 angstrom resolution. The DNA binds in the cleft and retains a B-DNA type of conformation. The enzyme, however, undergoes a series of conformational changes, including rotation of subunits and folding of disordered regions. The most striking conformational change is the unraveling of carboxyl-terminal alpha helices to form partially disordered "arms." The arm from one subunit fits into the minor groove while the arm from the symmetry related subunit follows the DNA sugar-phosphate backbone. Recognition of DNA base pairs occurs primarily in the major groove, with a few interactions occurring in the minor groove. Tightly bound water molecules play an equally important role as side chain and main chain atoms in the recognition of base pairs. The complex also provides new insights into the mechanism by which the enzyme catalyzes the hydrolysis of DNA phosphodiester groups.