Summary information and primary citation
- PDB-id
- 1bj6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-DNA
- Method
- NMR
- Summary
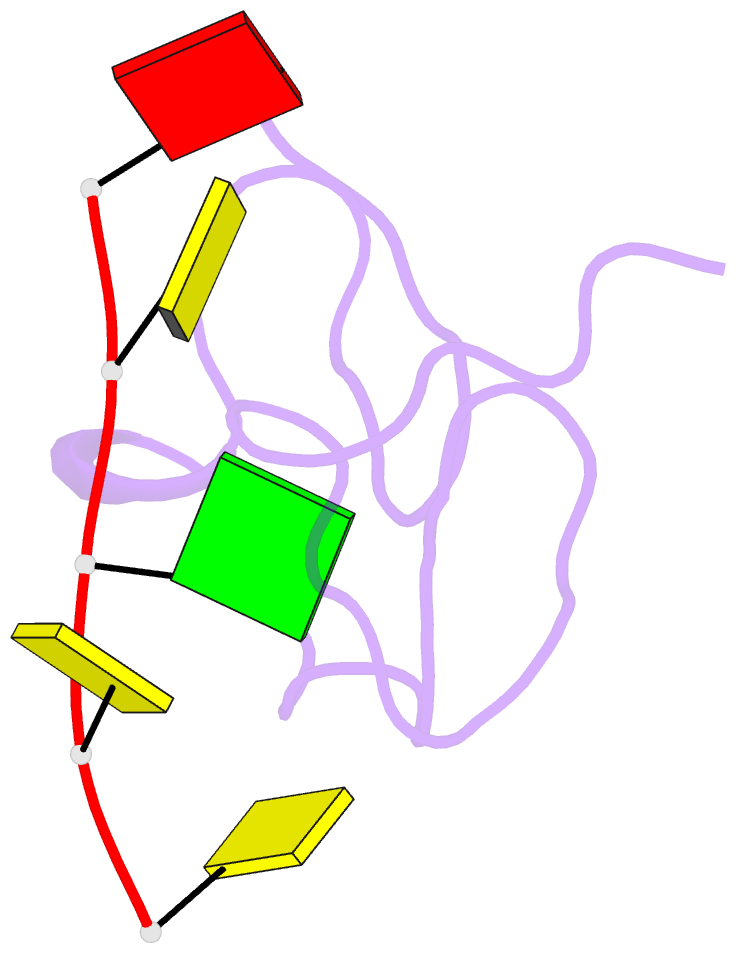
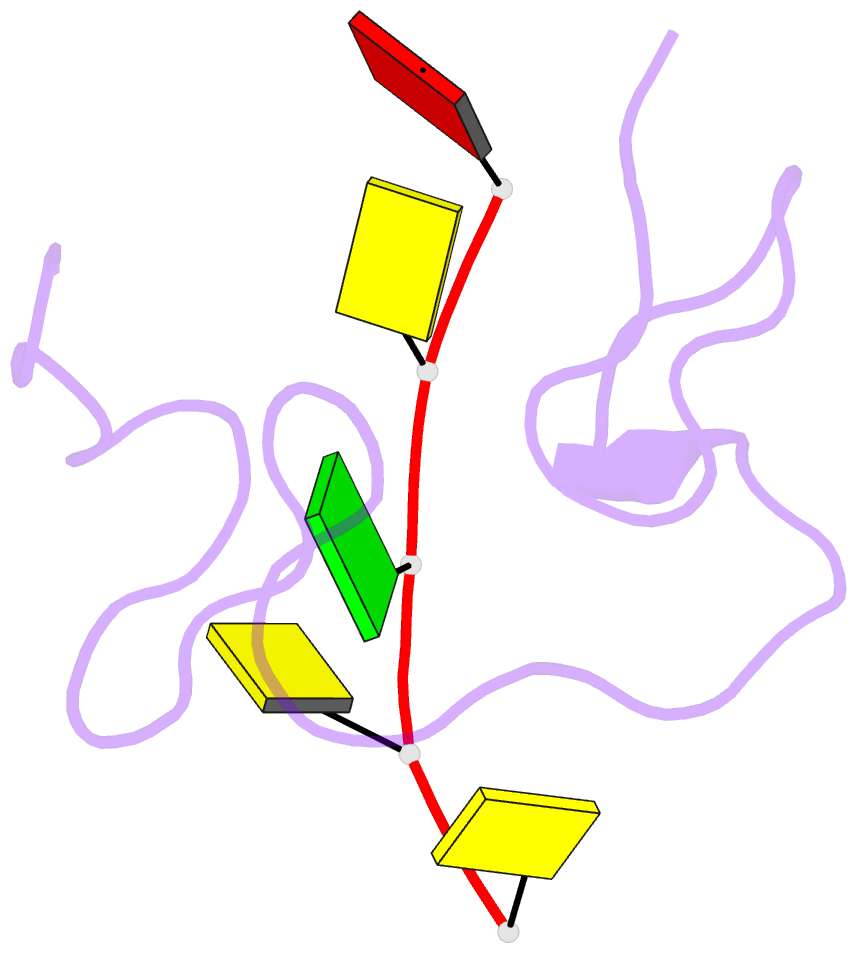
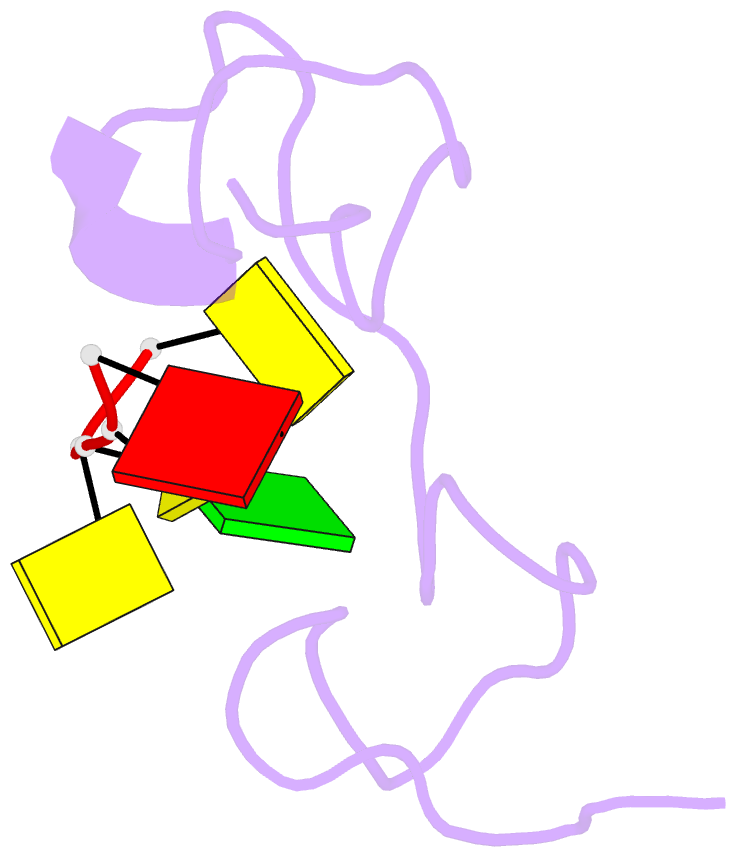
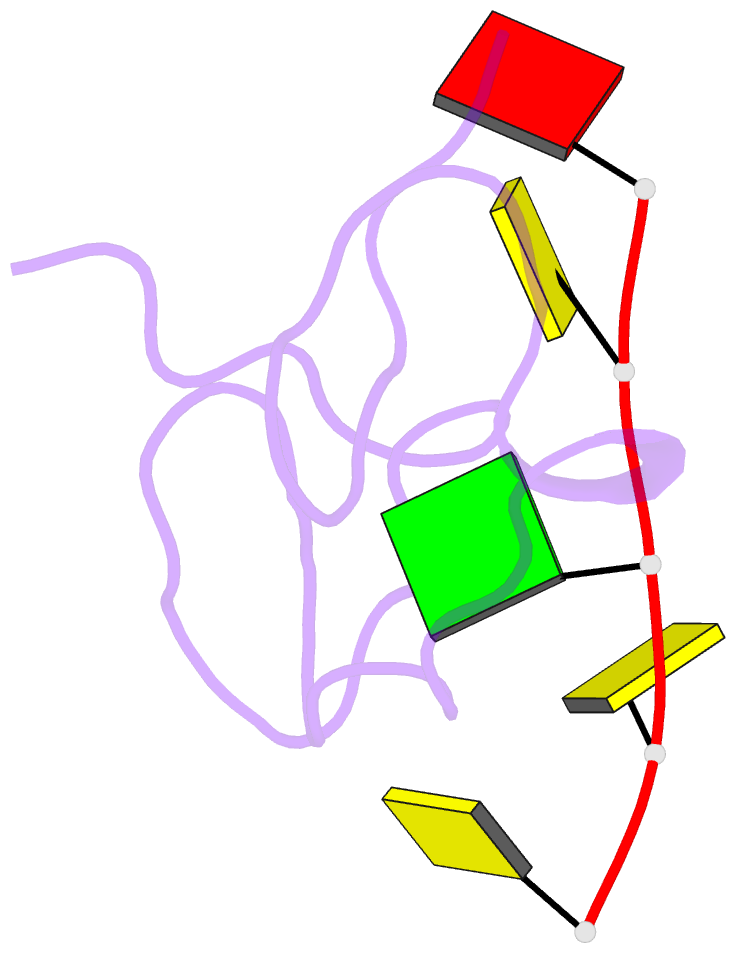
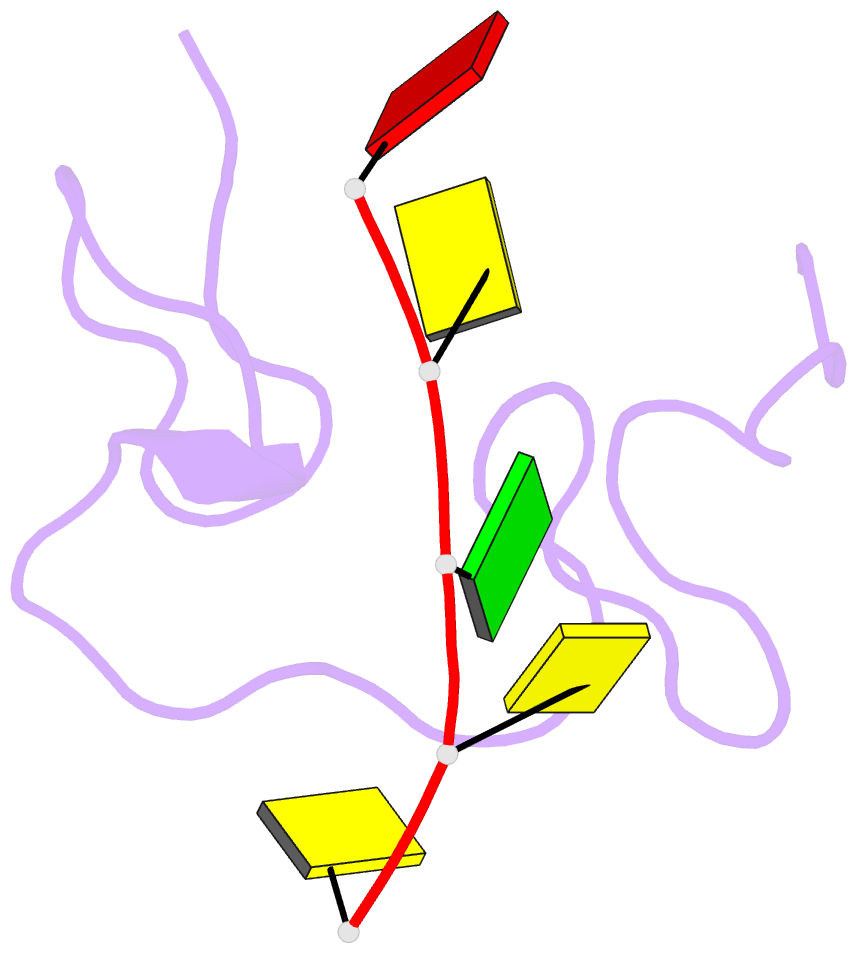
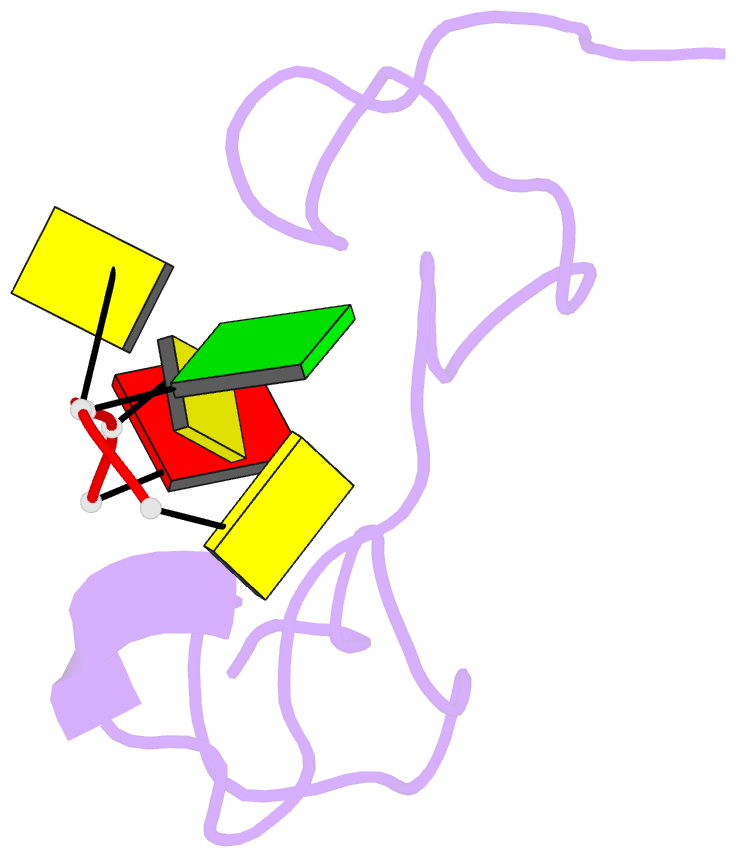
- 1h NMR of (12-53) ncp7-d(acgcc) complex, 10 structures
- Reference
- Morellet N, Demene H, Teilleux V, Huynh-Dinh T, de Rocquigny H, Fournie-Zaluski MC, Roques BP (1998): "Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC)." J.Mol.Biol., 283, 419-434. doi: 10.1006/jmbi.1998.2098.
- Abstract
- The nucleocapsid protein NCp7 of HIV-1 Mal contains two successive Zn knuckles of the CX2CX4HX4C type and plays a major role in virion morphogenesis, genomic RNA packaging and viral infectivity, mainly through single-stranded nucleic acid binding. We report here the study by 1H 2D NMR of the complex formed between the (12-53)NCp7, encompassing the two Zn knuckles, and d(ACGCC), a deoxynucleotide sequence analog corresponding to the shortest NCp7 binding site. Ten structures of the (12-53)NCp7/d(ACGCC) complex have been obtained from 607 NOE-derived distance constraints, 28 of which are intermolecular, and from molecular dynamics studies. The oligonucleotide is almost perpendicular to the sequence linking the two Zn knuckles. The Trp37 indole ring is inserted between the C2 and G3 bases and stacked on the latter. The complex is stabilized by hydrophobic interactions and hydrogen bonds, and accounts for the observed loss of virus infectivity induced by mutations in the Zn knuckle domain. Thus, the interaction between d(ACGCC) and the inactive mutant Cys23 (12-53)NCp7 was found by NMR to be completely different from that observed with the wild-type peptide. A mechanism of action for NCp7 in virus morphogenesis and replication is proposed from these results, which could facilitate the design of possible antiviral agents acting by a new mechanism.