Summary information and primary citation
- PDB-id
- 1bp7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.0 Å)
- Summary
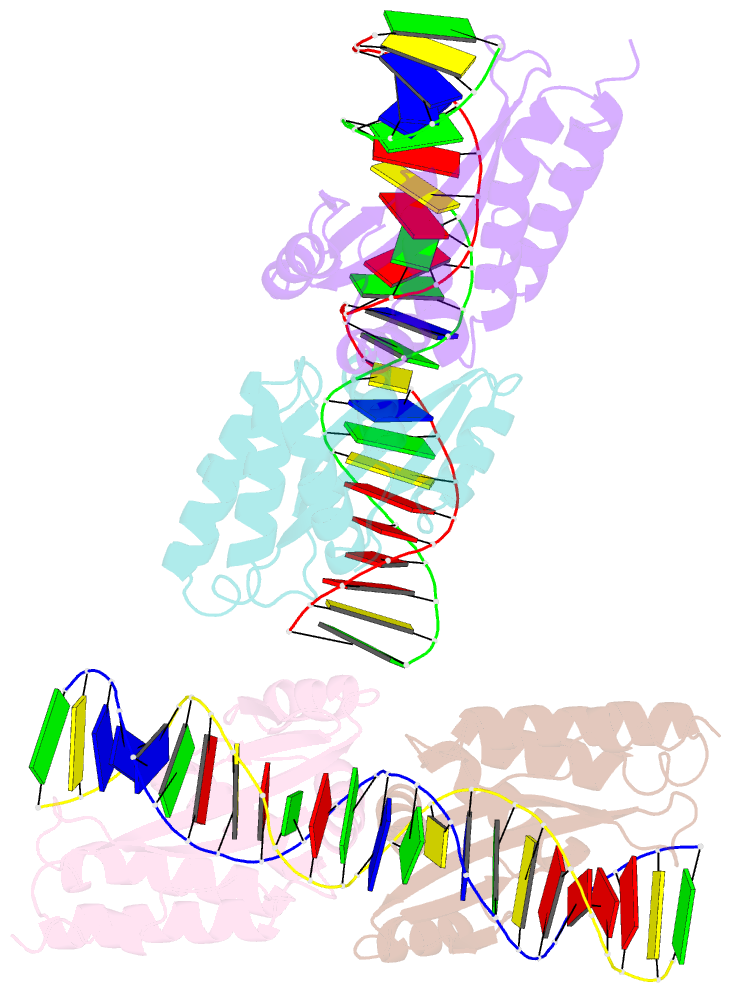
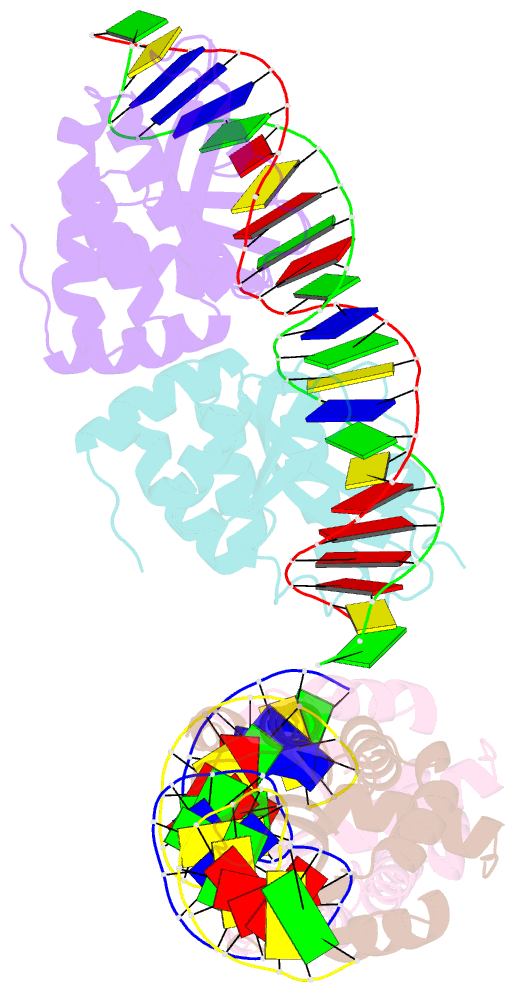
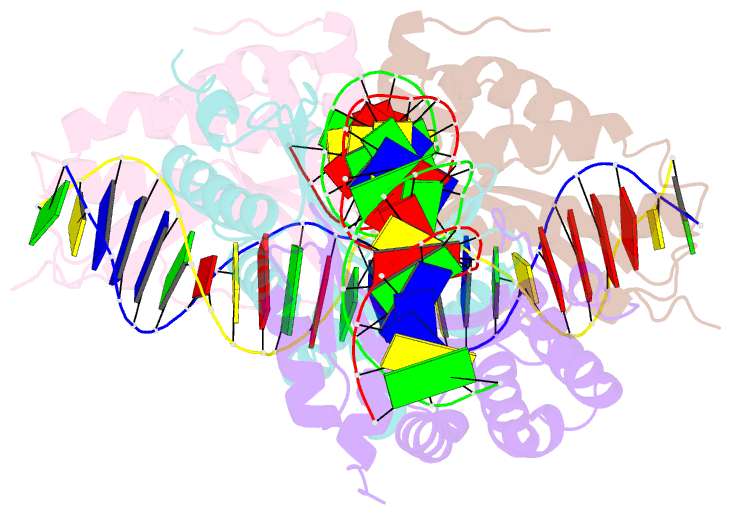
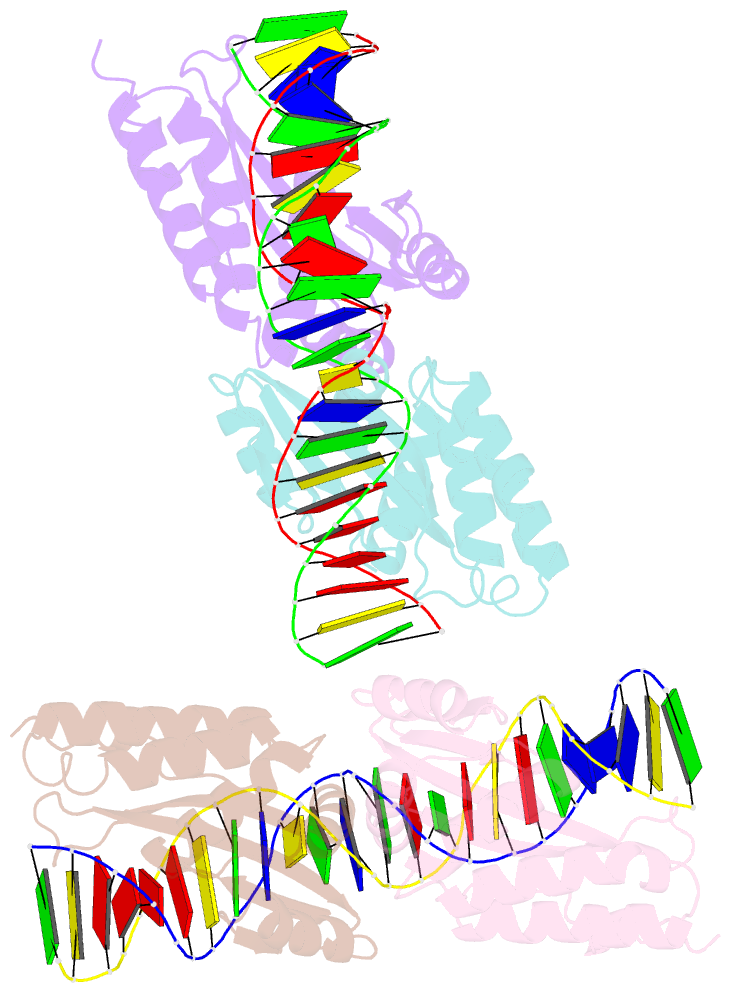
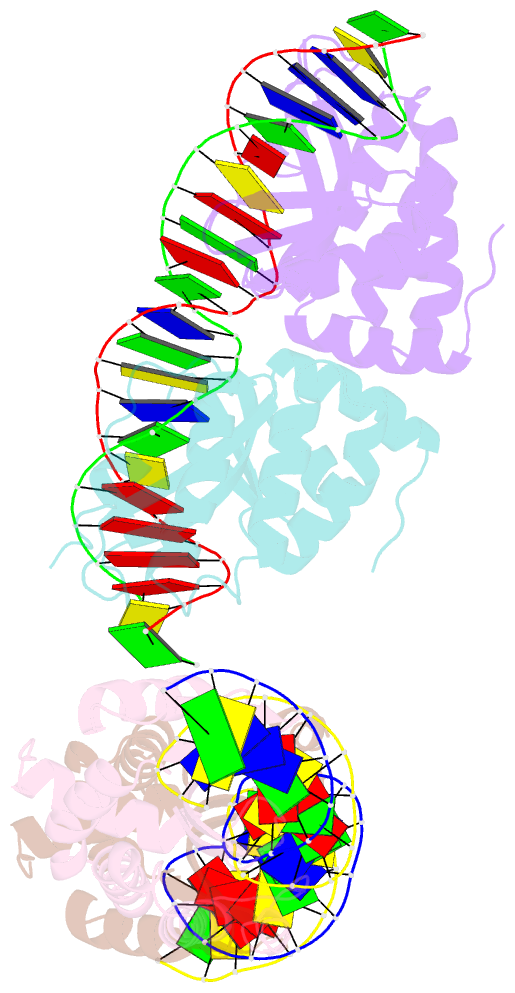
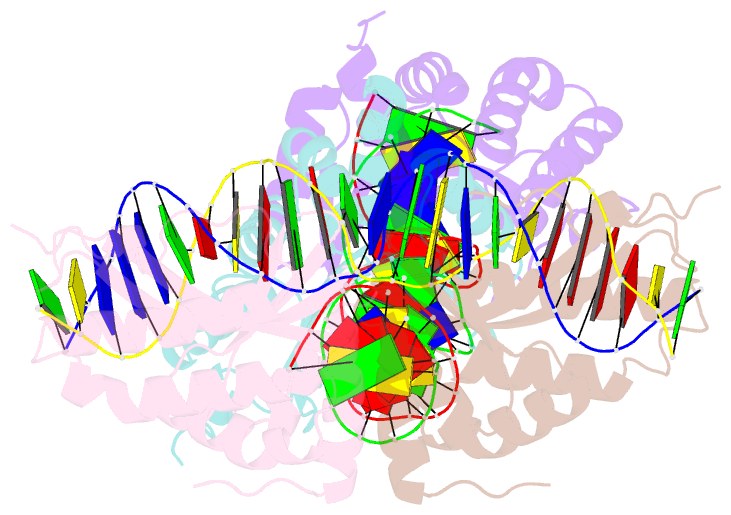
- Group i mobile intron endonuclease i-crei complexed with homing site DNA
- Reference
- Jurica MS, Monnat Jr RJ, Stoddard BL (1998): "DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI." Mol.Cell, 2, 469-476. doi: 10.1016/S1097-2765(00)80146-X.
- Abstract
- The structure of the LAGLIDADG intron-encoded homing endonuclease I-CreI bound to homing site DNA has been determined. The interface is formed by an extended, concave beta sheet from each enzyme monomer that contacts each DNA half-site, resulting in direct side-chain contacts to 18 of the 24 base pairs across the full-length homing site. The structure indicates that I-CreI is optimized to its role in genetic transposition by exhibiting long site-recognition while being able to cleave many closely related target sequences. DNA cleavage is mediated by a compact pair of active sites in the I-CreI homodimer, each of which contains a separate bound divalent cation.