Summary information and primary citation
- PDB-id
- 1c0a; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (2.4 Å)
- Summary
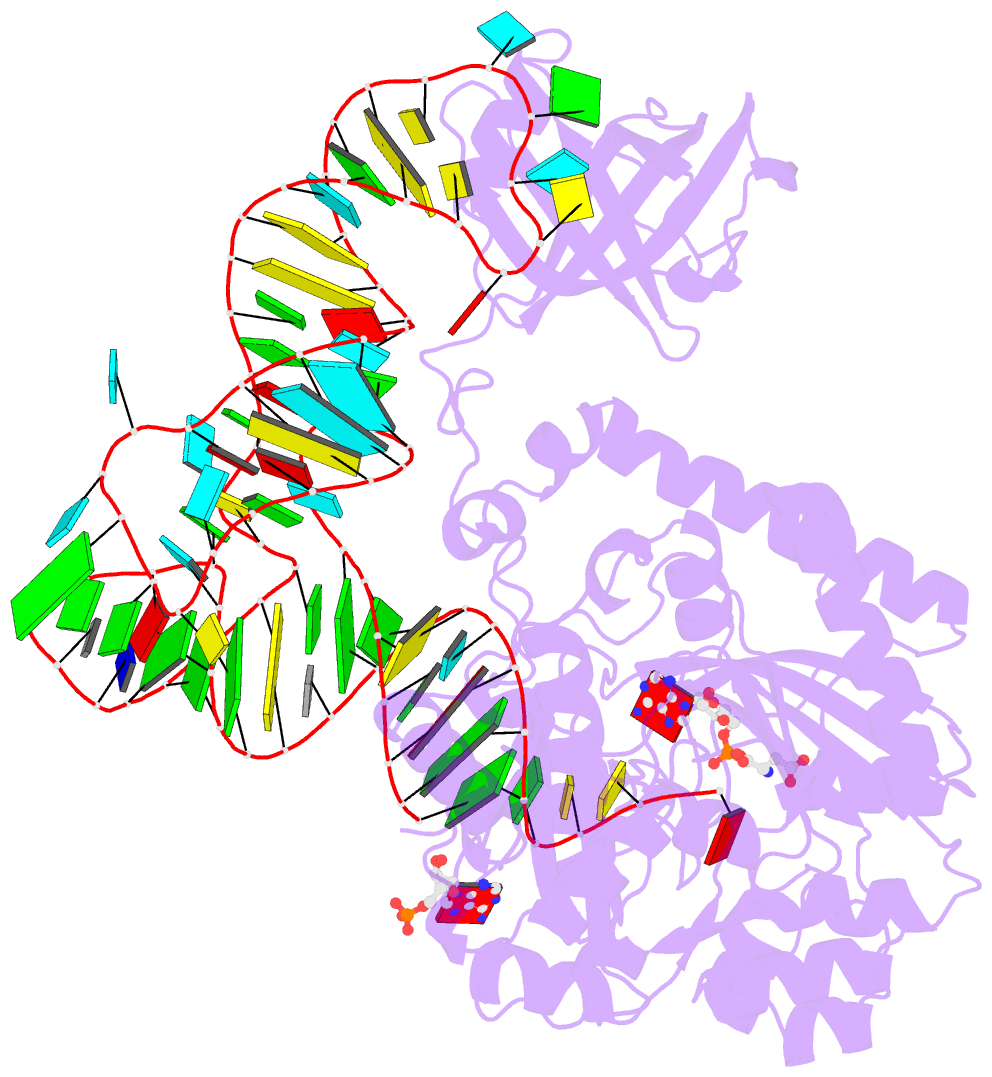
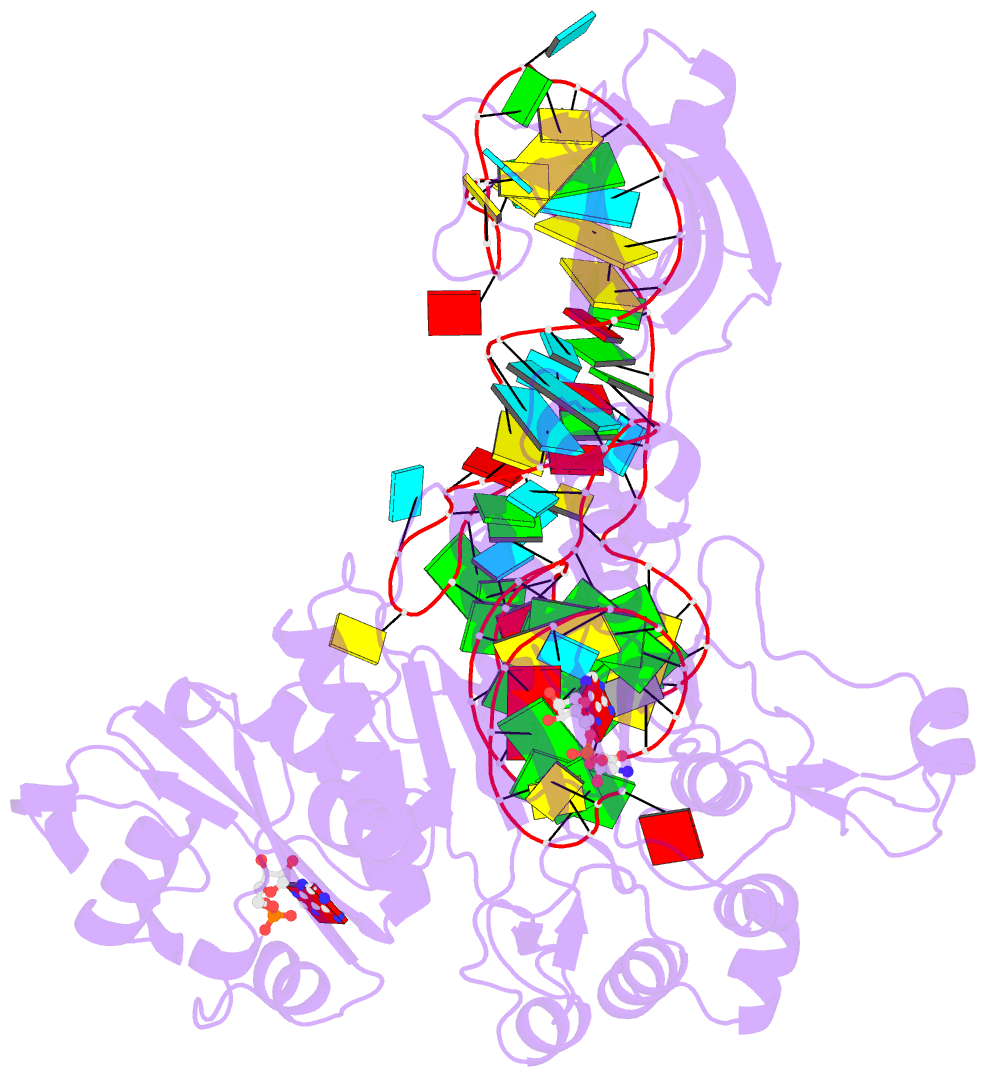
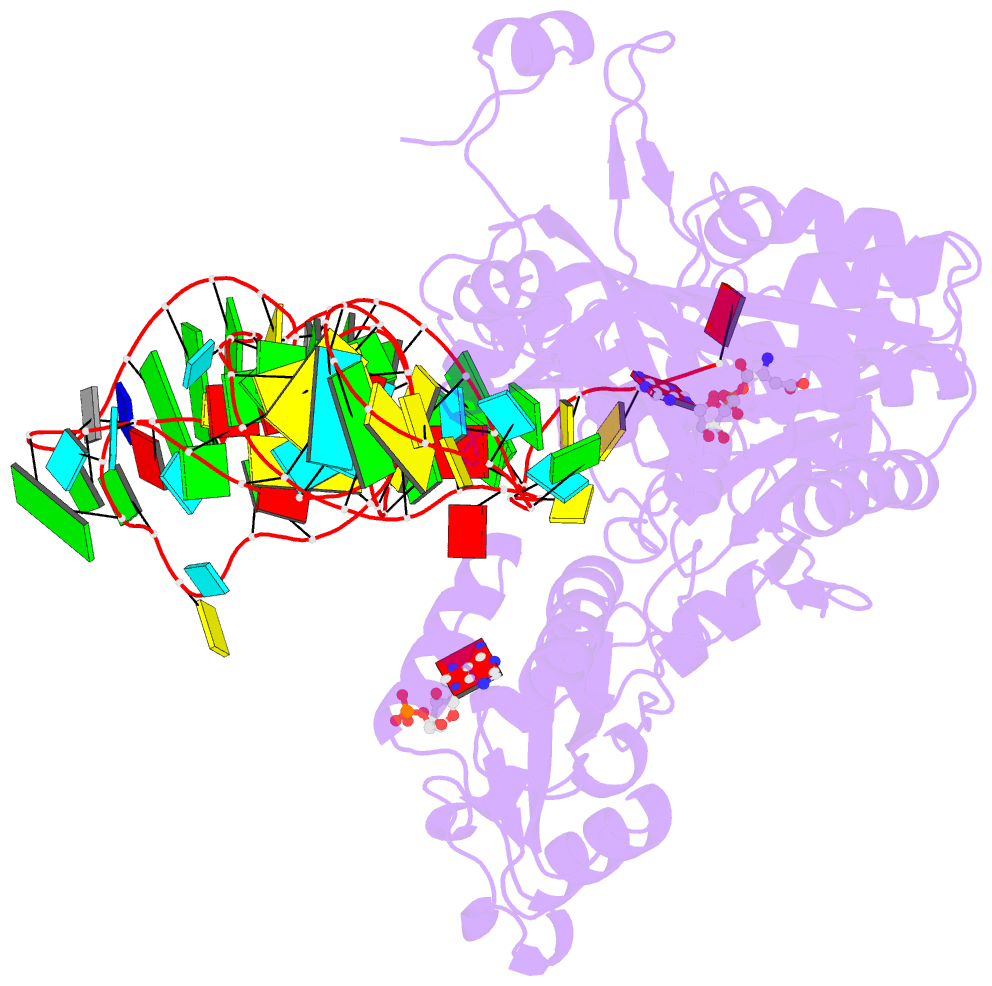
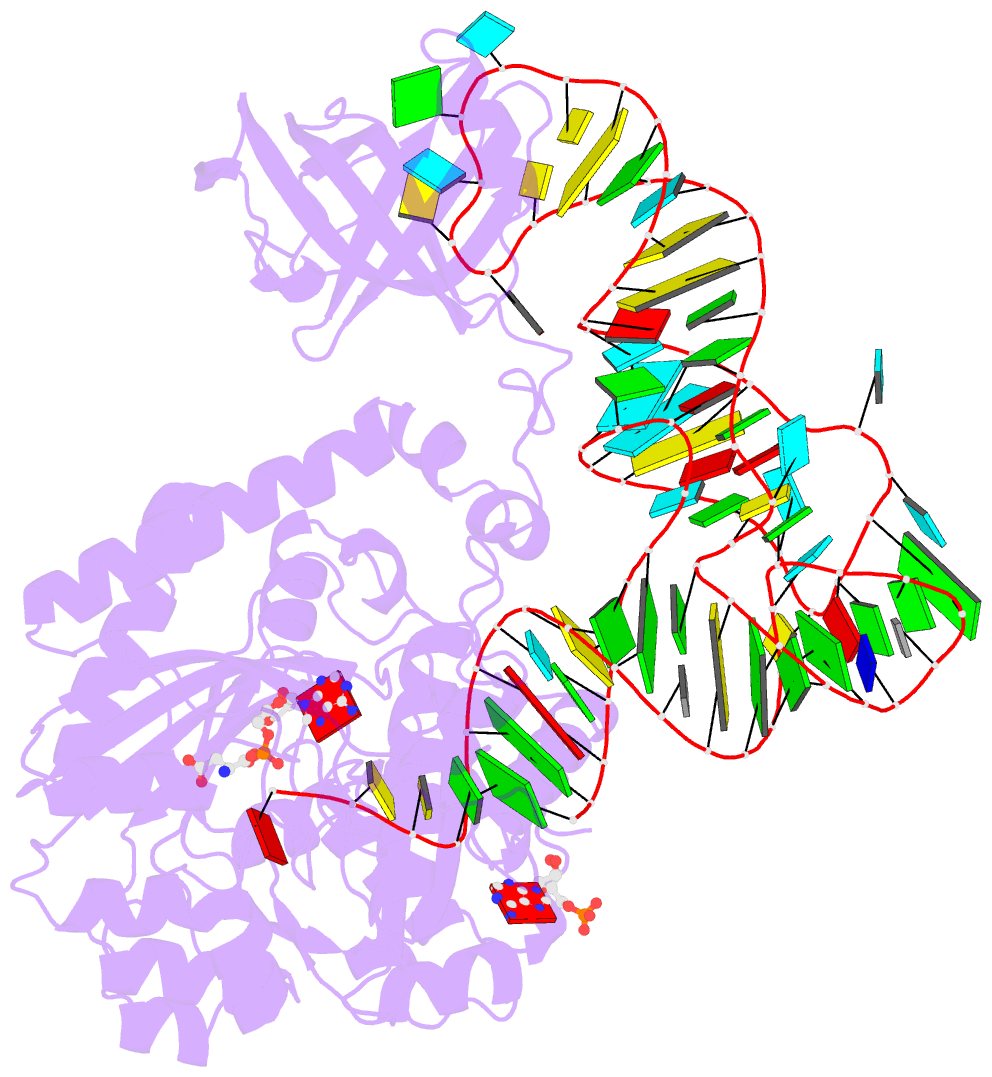
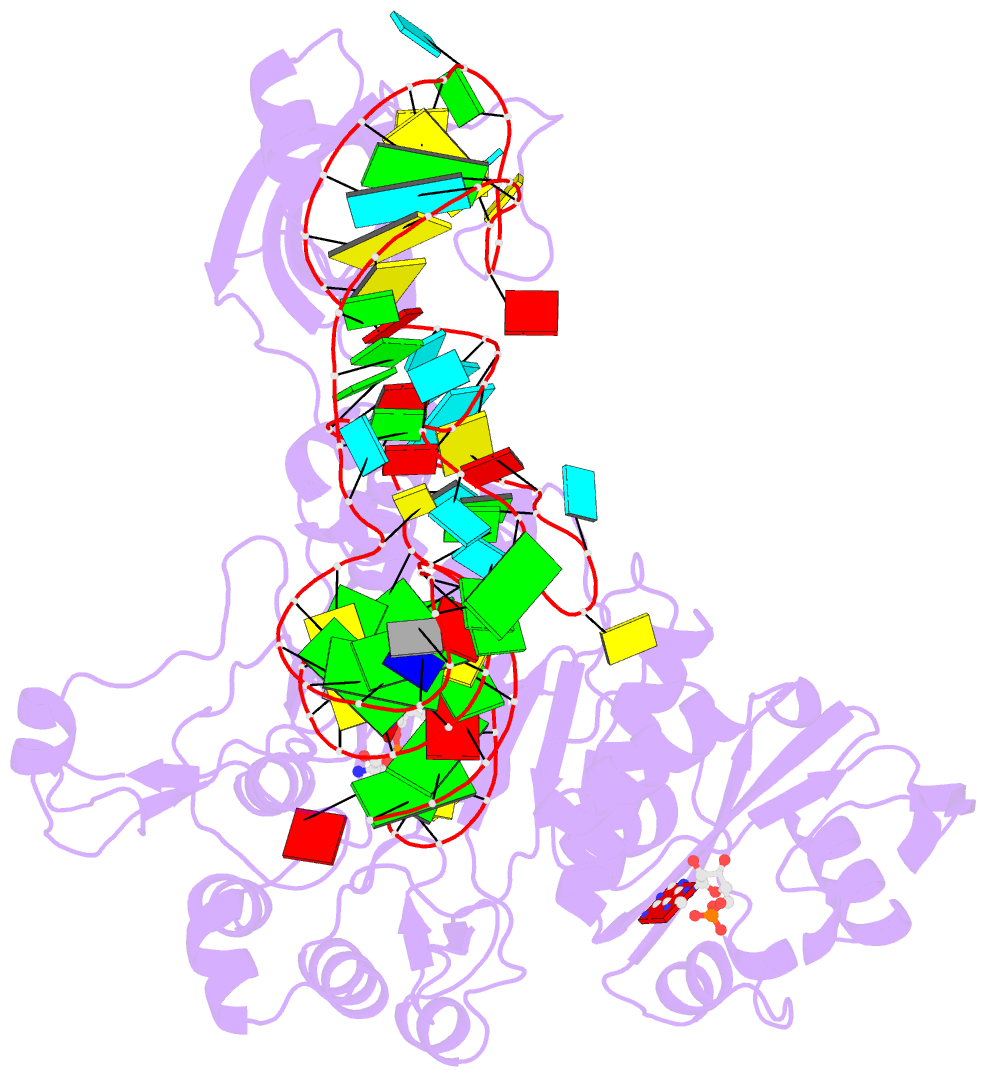
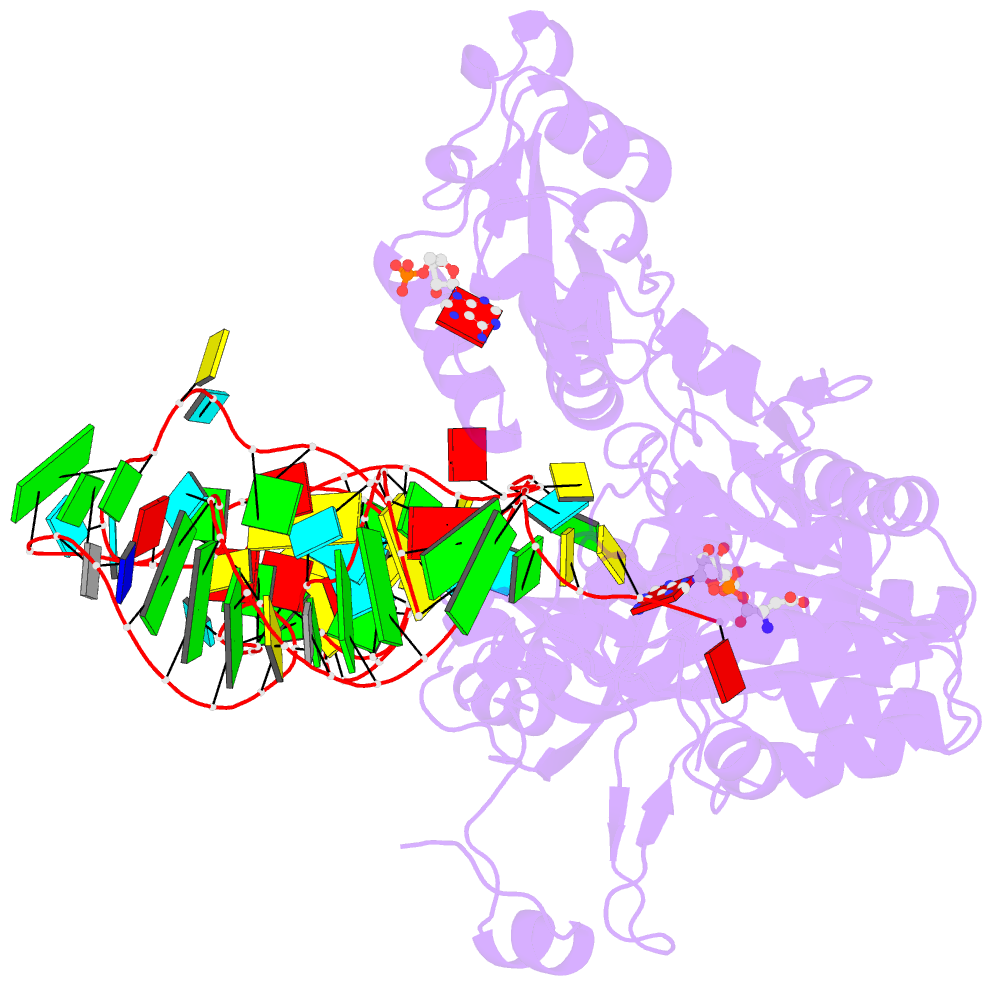
- Crystal structure of the e. coli aspartyl-trna synthetase : trnaasp : aspartyl-adenylate complex
- Reference
- Eiler S, Dock-Bregeon A, Moulinier L, Thierry JC, Moras D (1999): "Synthesis of aspartyl-tRNA(Asp) in Escherichia coli--a snapshot of the second step." EMBO J., 18, 6532-6541. doi: 10.1093/emboj/18.22.6532.
- Abstract
- The 2.4 A crystal structure of the Escherichia coli aspartyl-tRNA synthetase (AspRS)-tRNA(Asp)-aspartyl-adenylate complex shows the two substrates poised for the transfer of the aspartic acid moiety from the adenylate to the 3'-hydroxyl of the terminal adenosine of the tRNA. A general molecular mechanism is proposed for the second step of the aspartylation reaction that accounts for the observed conformational changes, notably in the active site pocket. The stabilization of the transition state is mediated essentially by two amino acids: the class II invariant arginine of motif 2 and the eubacterial-specific Gln231, which in eukaryotes and archaea is replaced by a structurally non-homologous serine. Two archetypal RNA-protein modes of interactions are observed: the anticodon stem-loop, including the wobble base Q, binds to the N-terminal beta-barrel domain through direct protein-RNA interactions, while the binding of the acceptor stem involves both direct and water-mediated hydrogen bonds in an original recognition scheme.