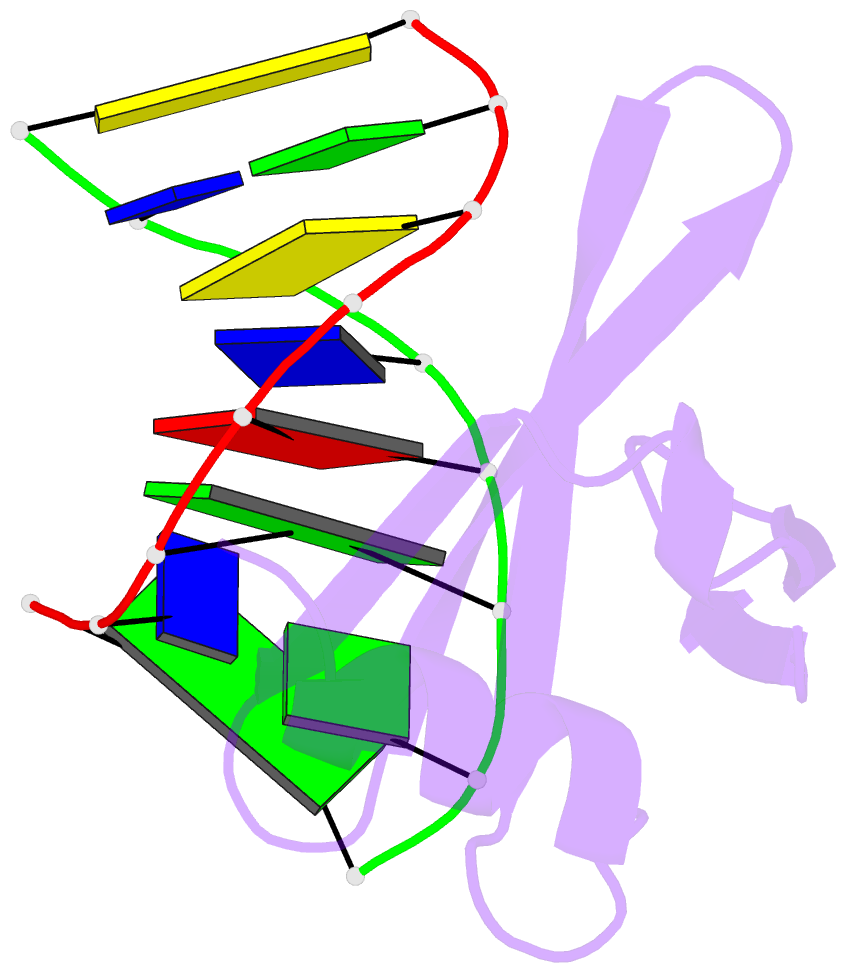
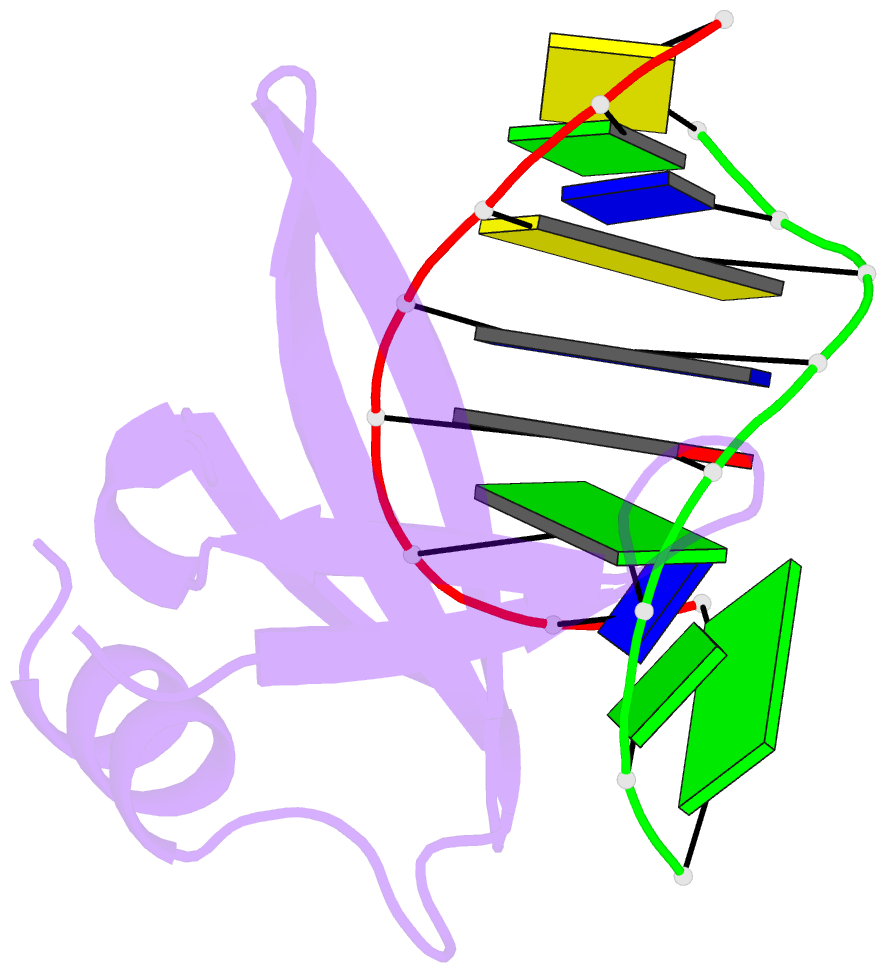
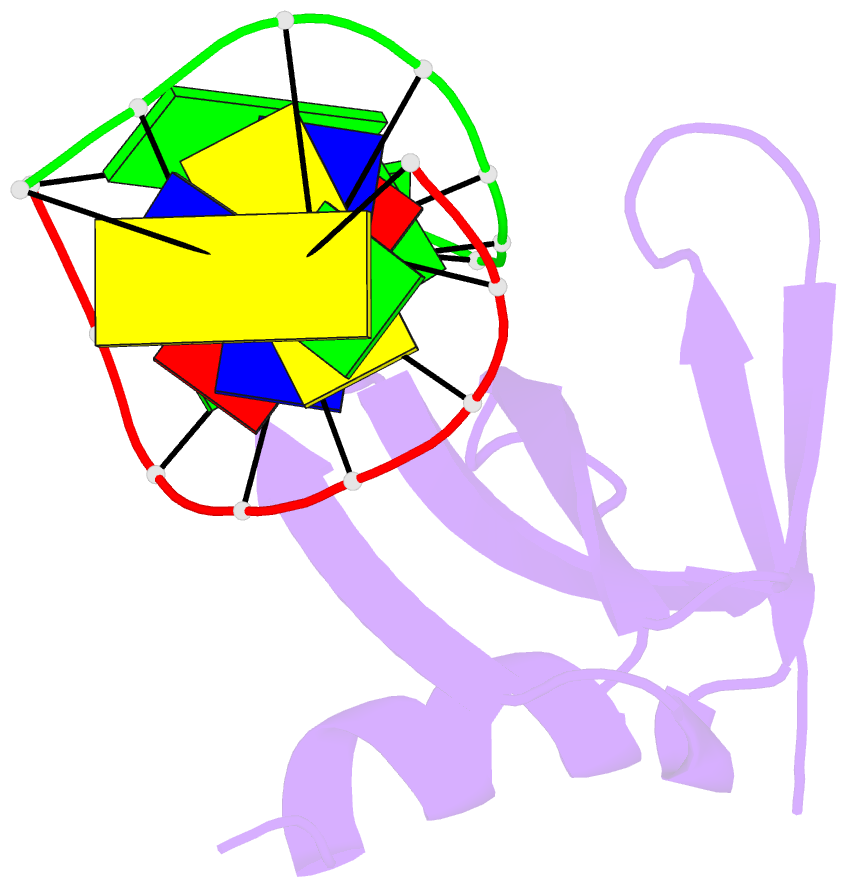
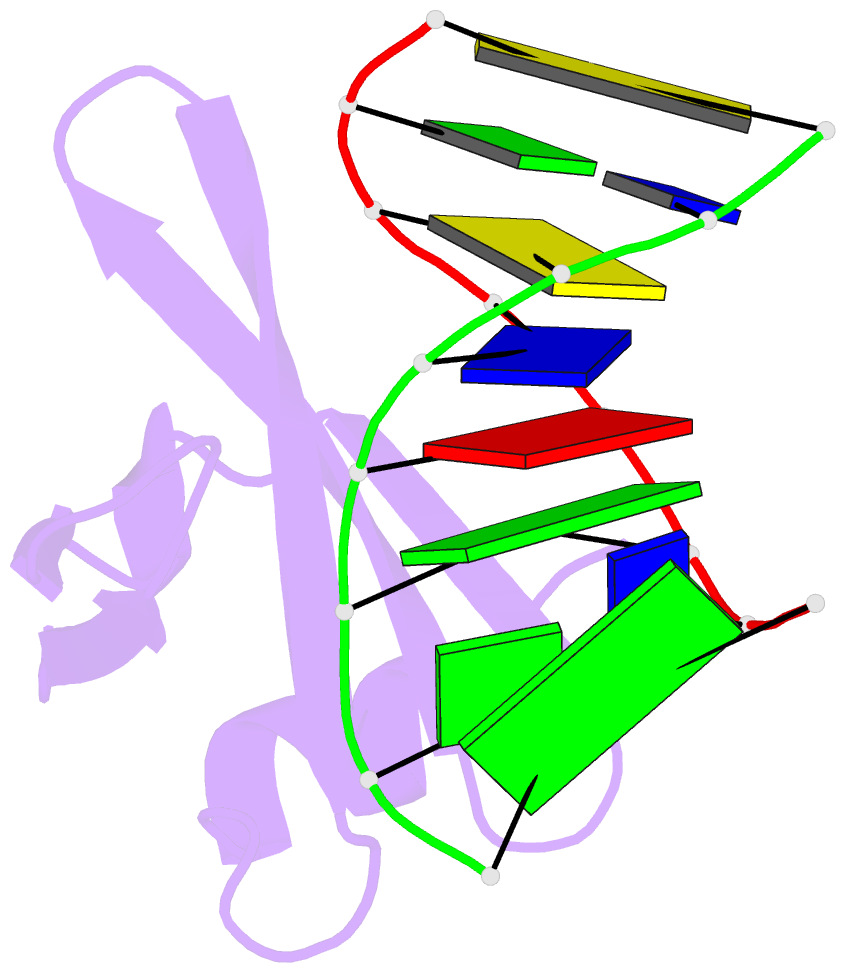
Summary information and primary citation
- PDB-id
- 1c8c; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.45 Å)
- Summary
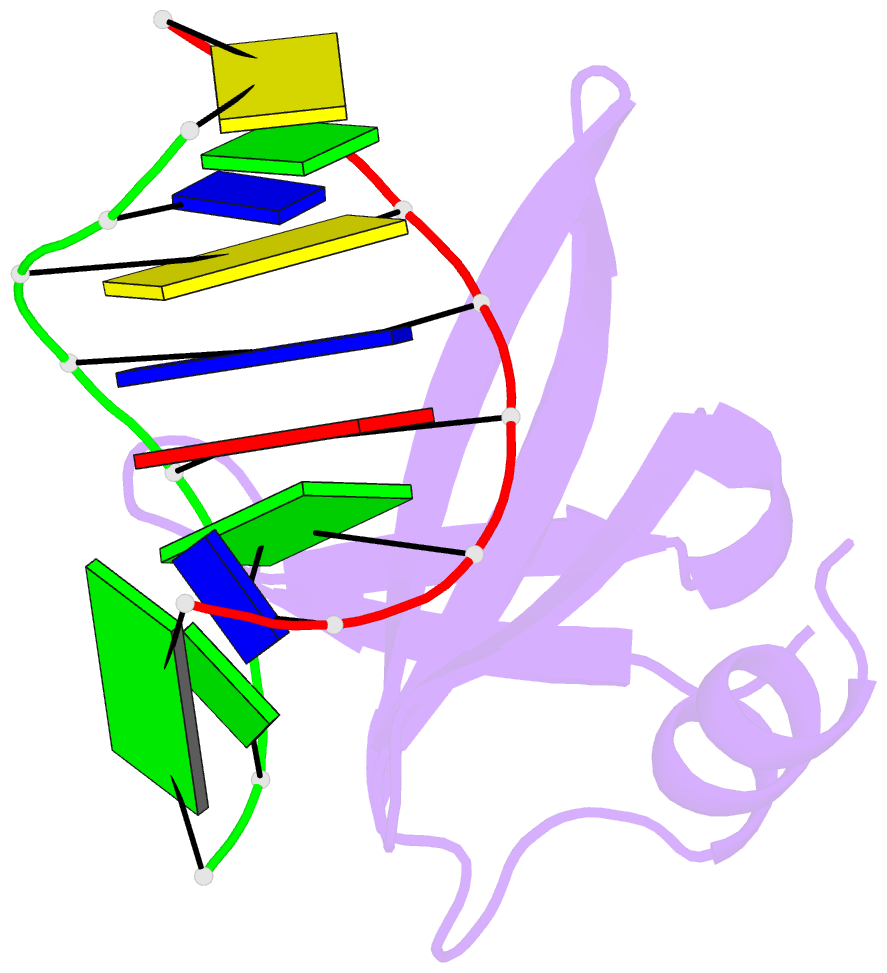
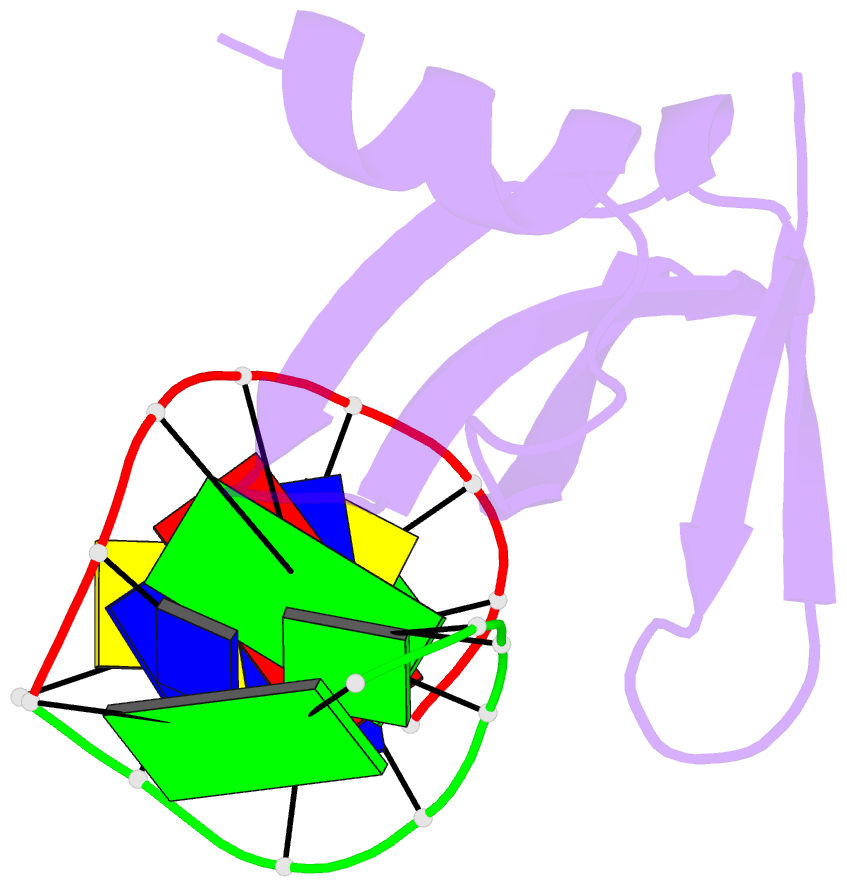
- Crystal structures of the chromosomal proteins sso7d-sac7d bound to DNA containing t-g mismatched base pairs
- Reference
- Su S, Gao YG, Robinson H, Liaw YC, Edmondson SP, Shriver JW, Wang AH (2000): "Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs." J.Mol.Biol., 303, 395-403. doi: 10.1006/jmbi.2000.4112.
- Abstract
- Sso7d and Sac7d are two small chromatin proteins from the hyperthermophilic archaeabacterium Sulfolobus solfataricus and Sulfolobus acidocaldarius, respectively. The crystal structures of Sso7d-GTGATCGC, Sac7d-GTGATCGC and Sac7d-GTGATCAC have been determined and refined at 1.45 A, 2.2 A and 2.2 A, respectively, to investigate the DNA binding property of Sso7d/Sac7d in the presence of a T-G mismatch base-pair. Detailed structural analysis revealed that the intercalation site includes the T-G mismatch base-pair and Sso7d/Sac7d bind to that mismatch base-pair in a manner similar to regular DNA. In the Sso7d-GTGATCGC complex, a new inter-strand hydrogen bond between T2O4 and C14N4 is formed and well-order bridging water molecules are found. The results suggest that the less stable DNA stacking site involving a T-G mismatch may be a preferred site for protein side-chain intercalation.