Summary information and primary citation
- PDB-id
- 1cez; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.4 Å)
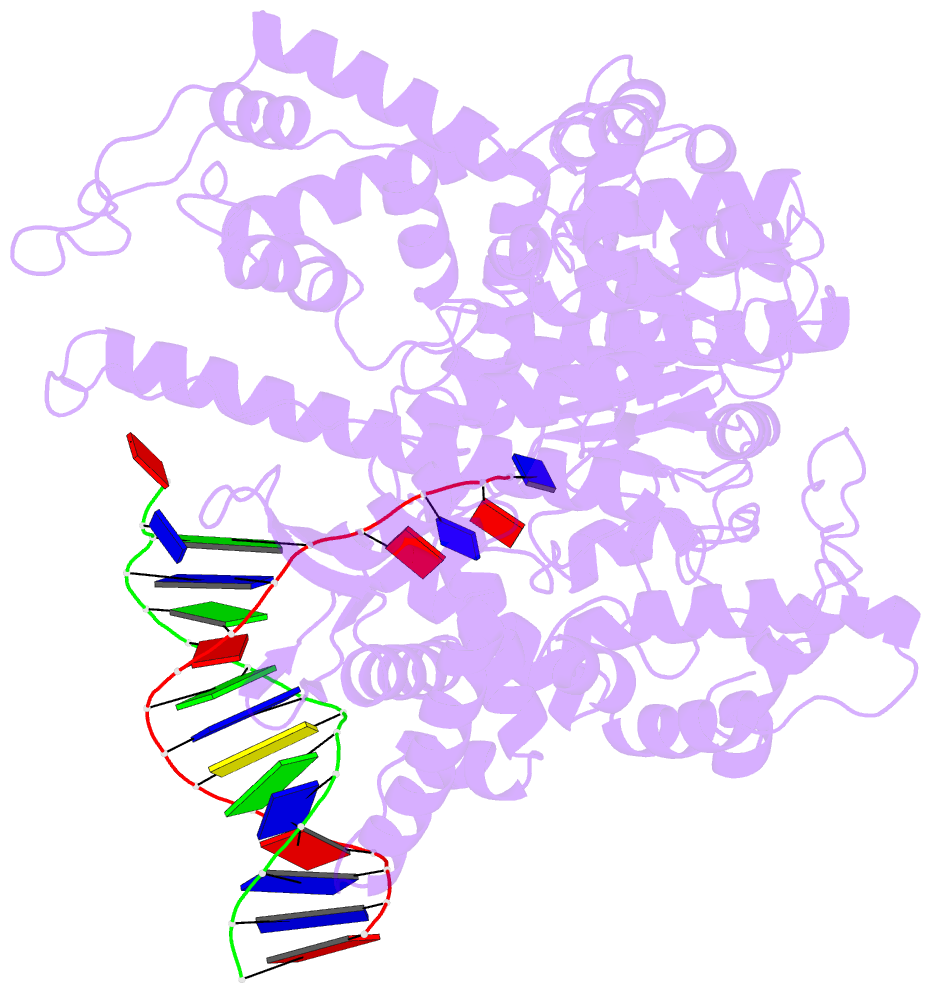
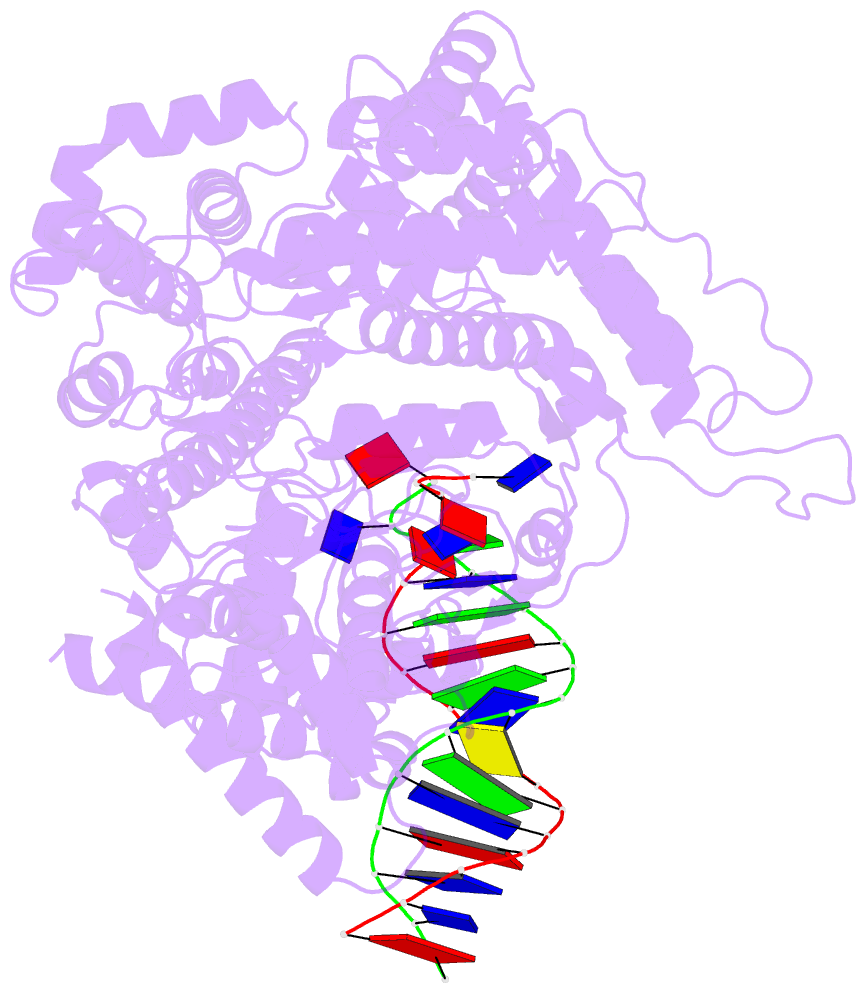
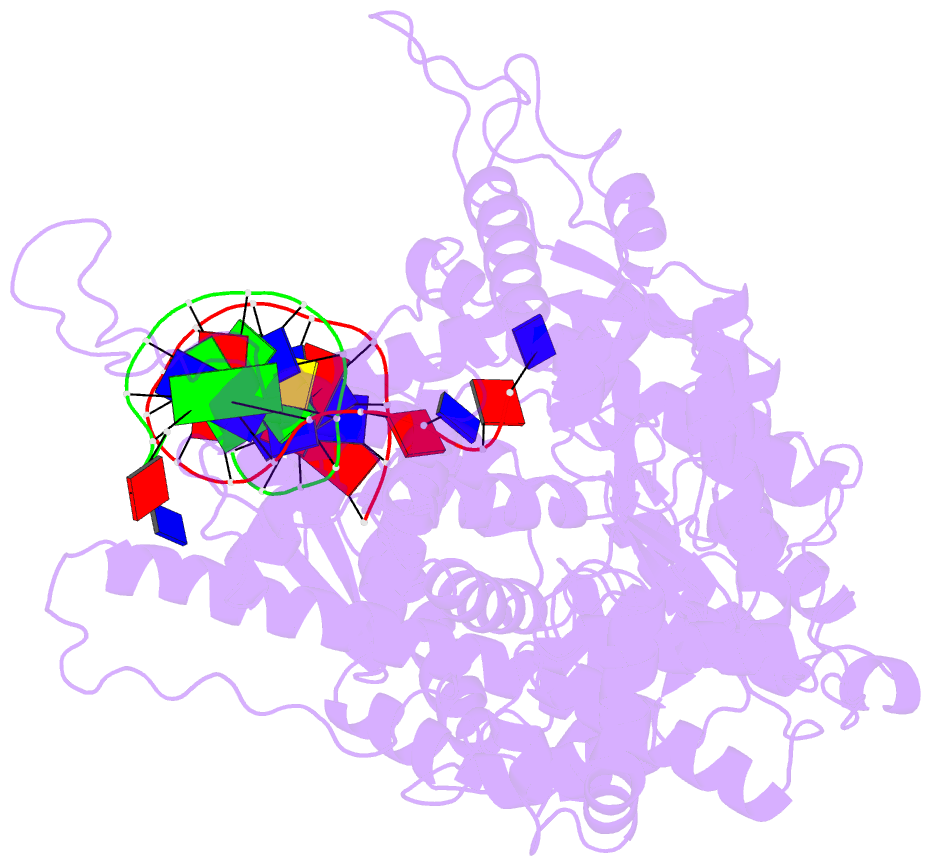
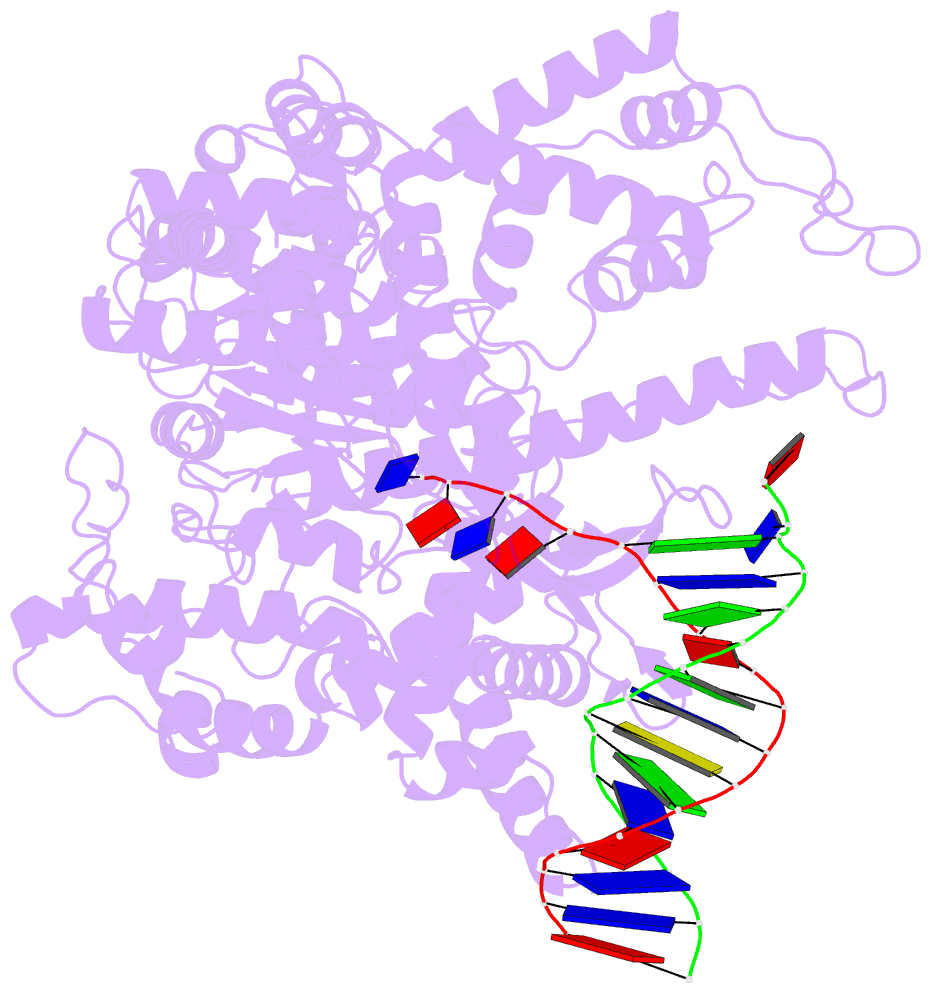
- Summary
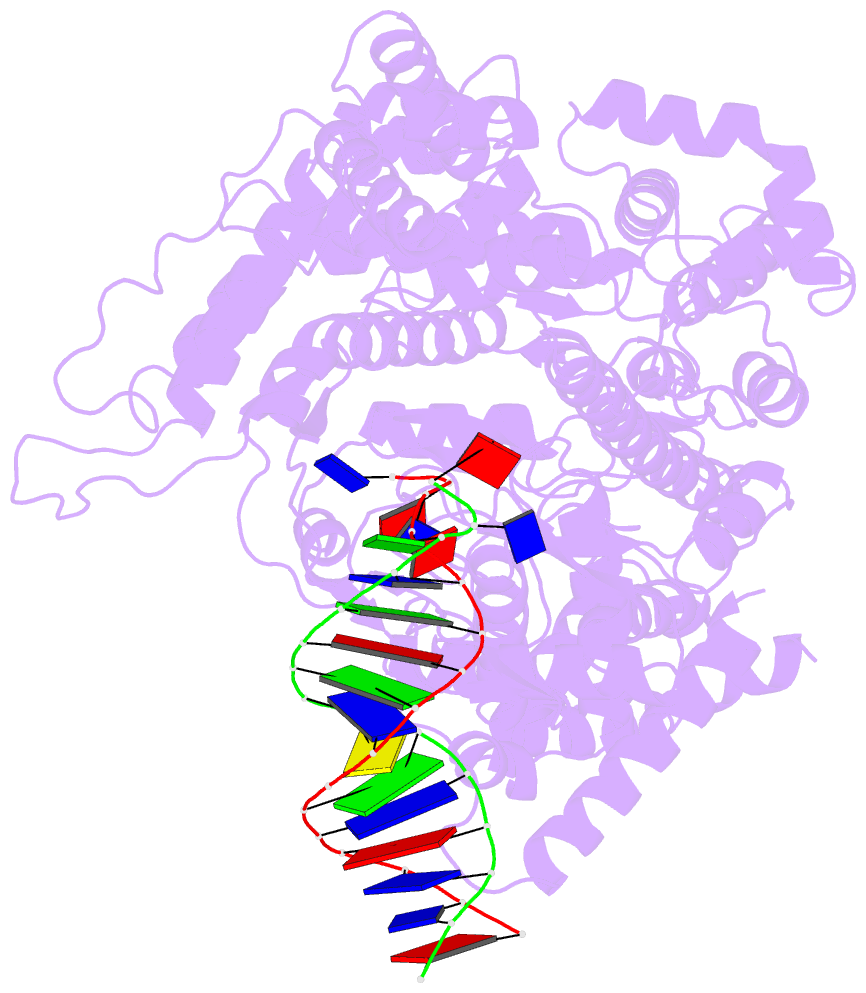
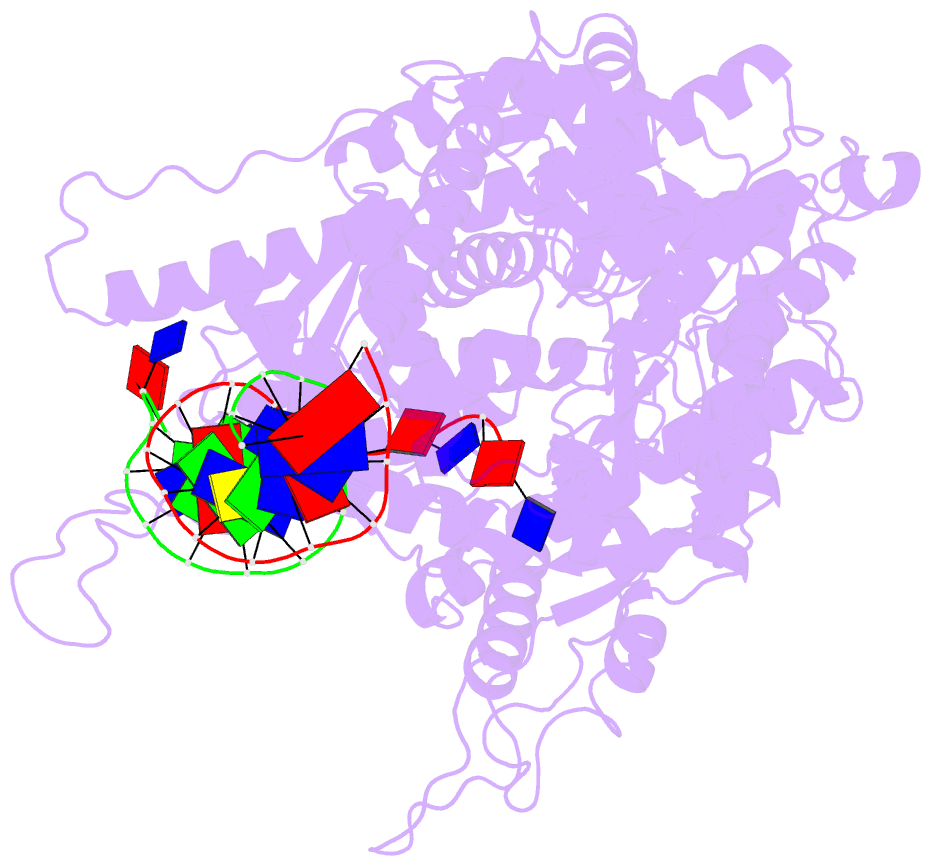
- Crystal structure of a t7 RNA polymerase-t7 promoter complex
- Reference
- Cheetham GM, Jeruzalmi D, Steitz TA (1999): "Structural basis for initiation of transcription from an RNA polymerase-promoter complex." Nature, 399, 80-83. doi: 10.1038/19999.
- Abstract
- Although the single-polypeptide-chain RNA polymerase from bacteriophage T7 (T7RNAP), like other RNA polymerases, uses the same mechanism of polymerization as the DNA polymerases, it can also recognize a specific promoter sequence, initiate new RNA chains from a single nucleotide, abortively cycle the synthesis of short transcripts, be regulated by a transcription inhibitor, and terminate transcription. As T7RNAP is homologous to the Pol I family of DNA polymerases, the differences between the structure of T7RNAP complexed to substrates and that of the corresponding DNA polymerase complex provides a structural basis for understanding many of these functional differences. T7RNAP initiates RNA synthesis at promoter sequences that are conserved from positions -17 to +6 relative to the start site of transcription. The crystal structure at 2.4 A resolution of T7RNAP complexed with a 17-base-pair promoter shows that the four base pairs closest to the catalytic active site have melted to form a transcription bubble. The T7 promoter sequence is recognized by interactions in the major groove between an antiparallel beta-loop and bases. The amino-terminal domain is involved in promoter recognition and DNA melting. We have also used homology modelling of the priming and incoming nucleoside triphosphates from the T7 DNA-polymerase ternary complex structure to explain the specificity of T7RNAP for ribonucleotides, its ability to initiate from a single nucleotide, and the abortive cycling at the initiation of transcription.