Summary information and primary citation
- PDB-id
- 1ckt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation-DNA
- Method
- X-ray (2.5 Å)
- Summary
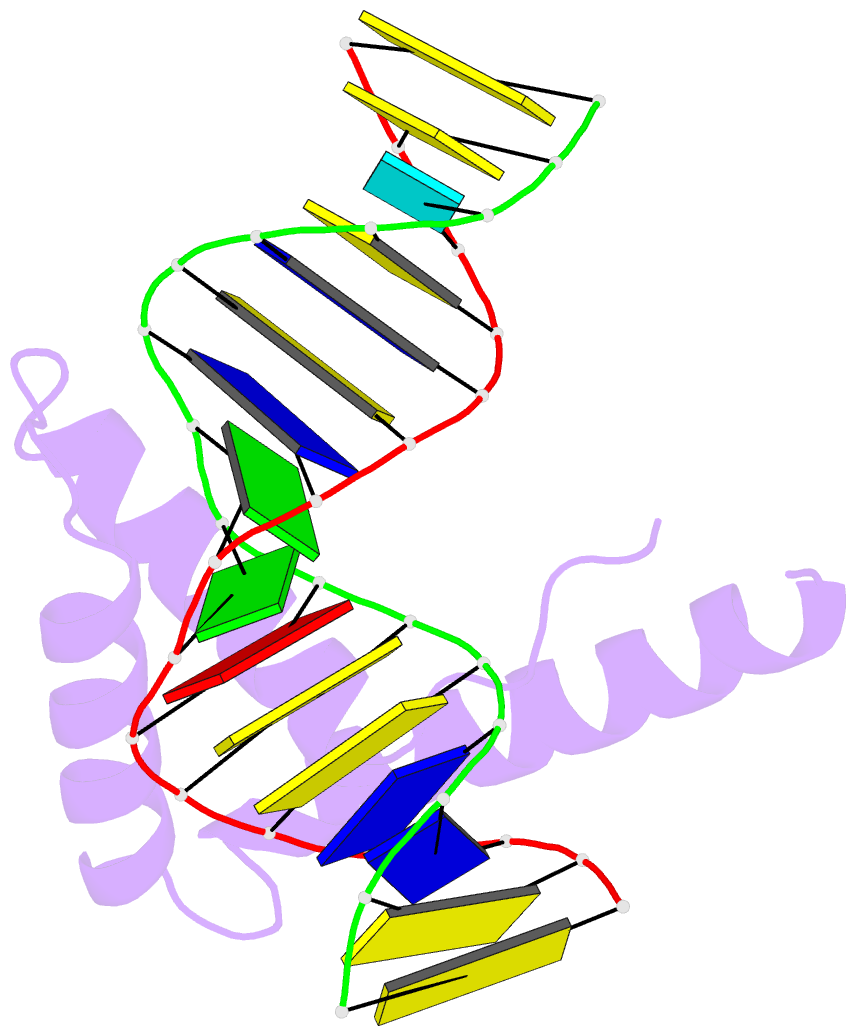
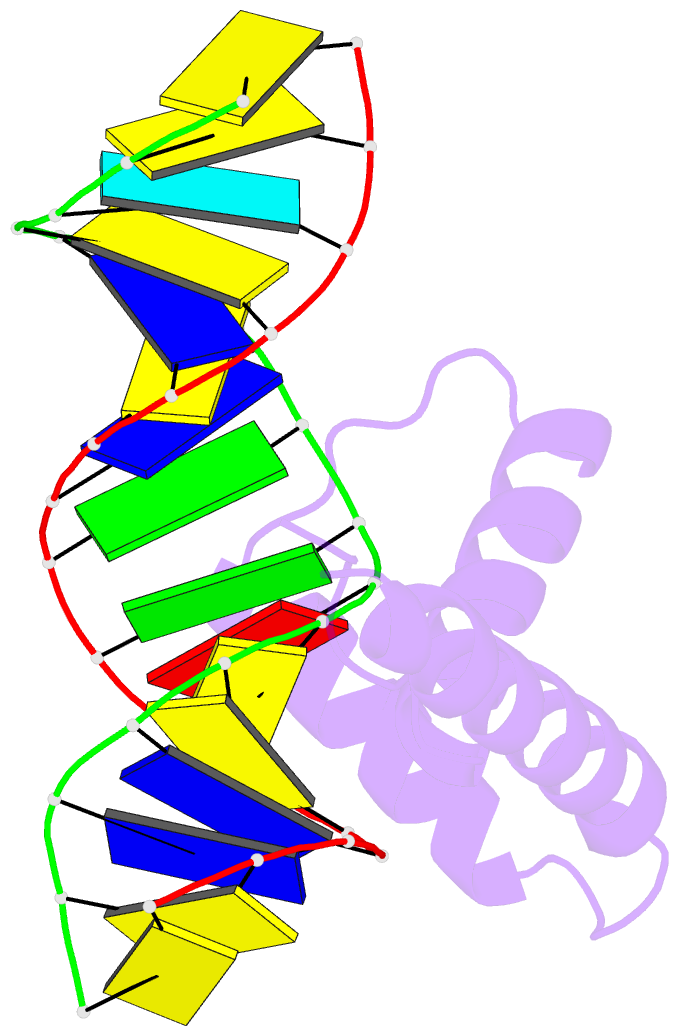
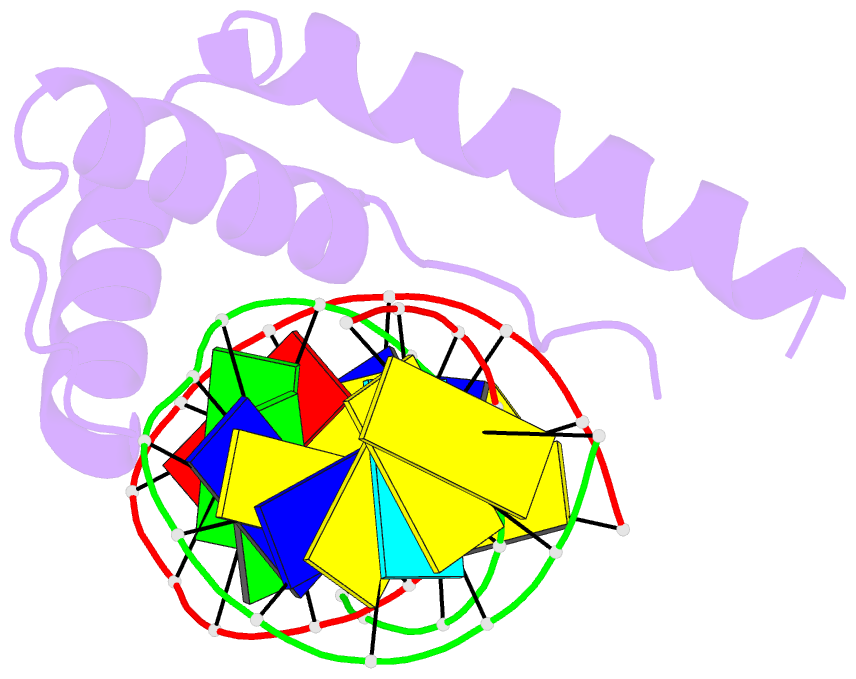
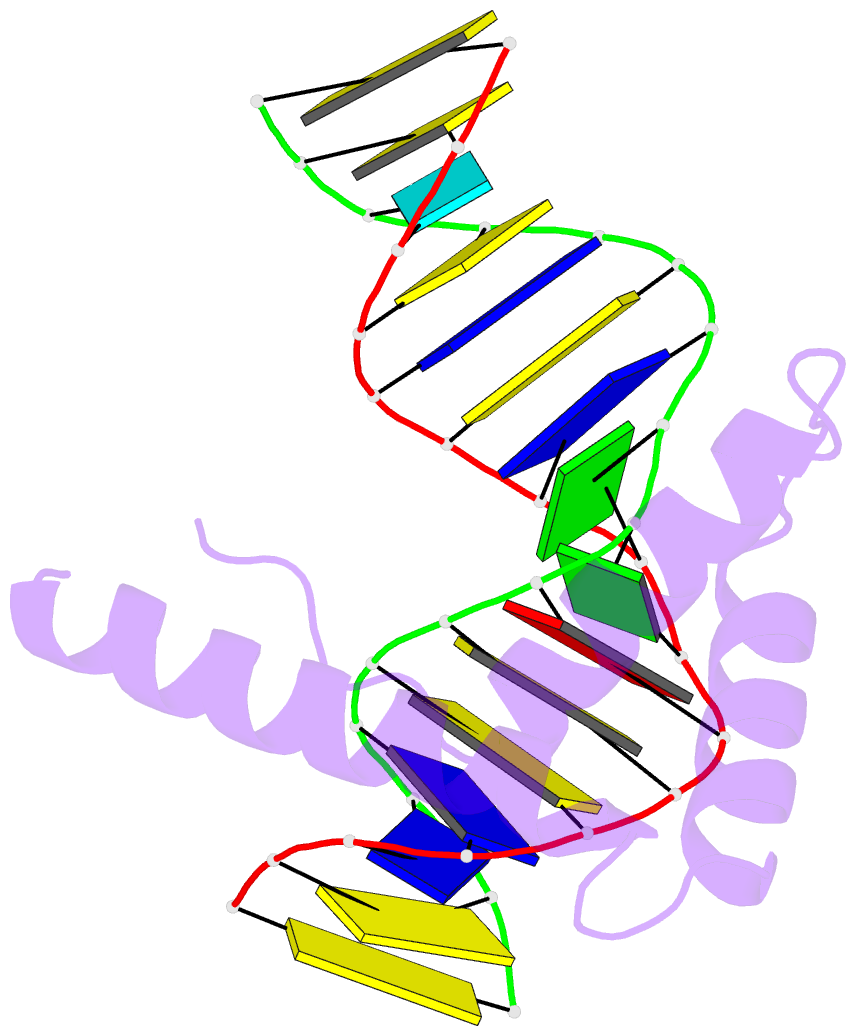
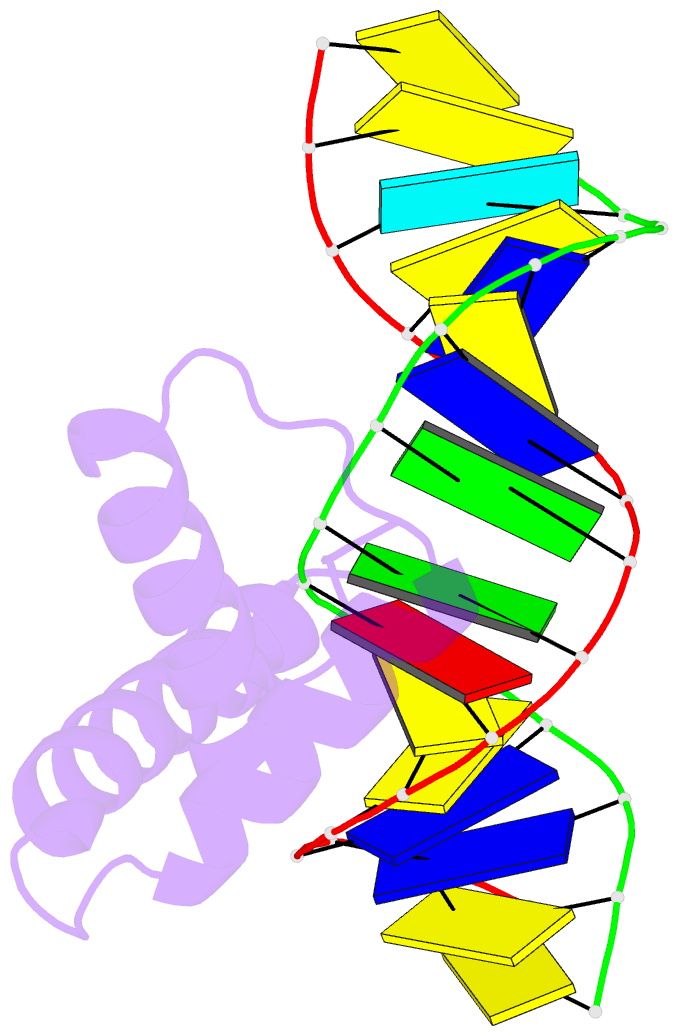
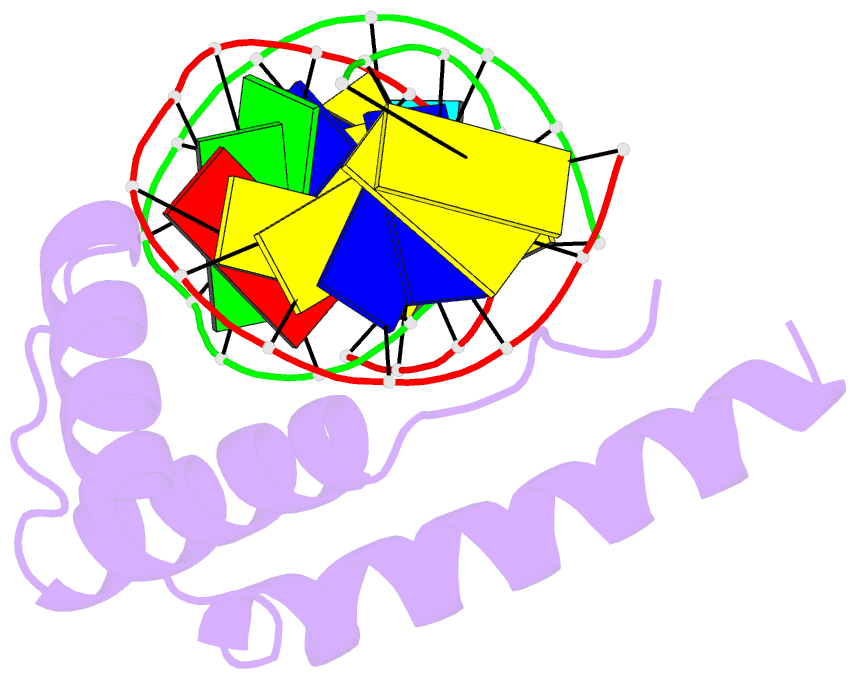
- Crystal structure of hmg1 domain a bound to a cisplatin-modified DNA duplex
- Reference
- Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ (1999): "Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins." Nature, 399, 708-712. doi: 10.1038/21460.
- Abstract
- The anticancer activity of cis-diamminedichloroplatinum(II) (cisplatin) arises from its ability to damage DNA, with the major adducts formed being intrastrand d(GpG) and d(ApG) crosslinks. These crosslinks bend and unwind the duplex, and the altered structure attracts high-mobility-group domain (HMG) and other proteins. This binding of HMG-domain proteins to cisplatin-modified DNA has been postulated to mediate the antitumour properties of the drug. Many HMG-domain proteins recognize altered DNA structures such as four-way junctions and cisplatin-modified DNA, but until now the molecular basis for this recognition was unknown. Here we describe mutagenesis, hydroxyl-radical footprinting and X-ray studies that elucidate the structure of a 1:1 cisplatin-modified DNA/HMG-domain complex. Domain A of the structure-specific HMG-domain protein HMG1 binds to the widened minor groove of a 16-base-pair DNA duplex containing a site-specific cis-[Pt(NH3)2[d(GpG)-N7(1),-N7(2)]] adduct. The DNA is strongly kinked at a hydrophobic notch created at the platinum-DNA crosslink and protein binding extends exclusively to the 3' side of the platinated strand. A phenylalanine residue at position 37 intercalates into a hydrophobic notch created at the platinum crosslinked d(GpG) site and binding of the domain is dramatically reduced in a mutant in which alanine is substituted for phenylalanine at this position.