Summary information and primary citation
- PDB-id
- 1d02; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.7 Å)
- Summary
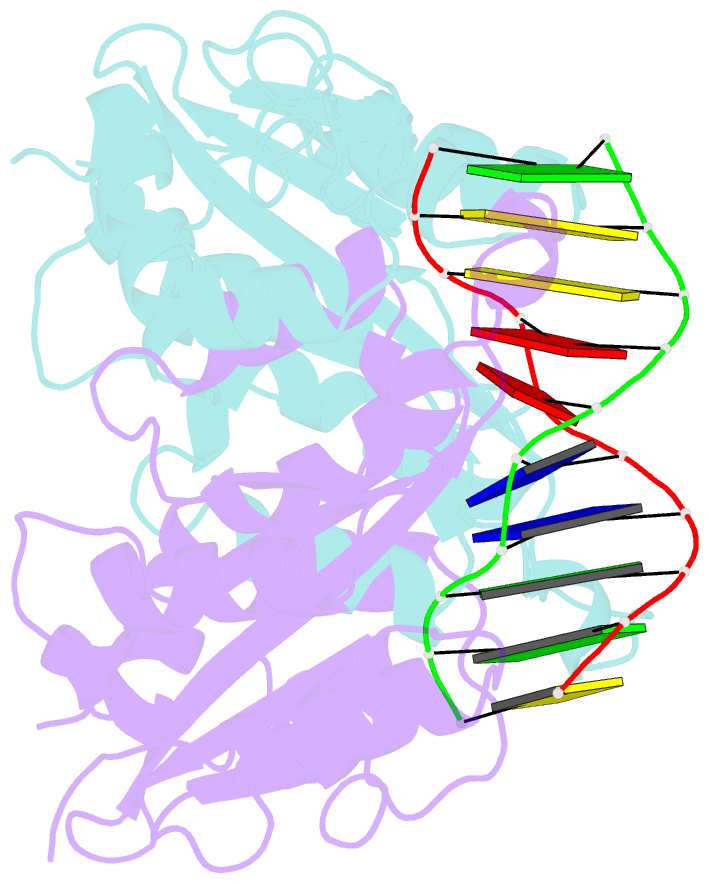
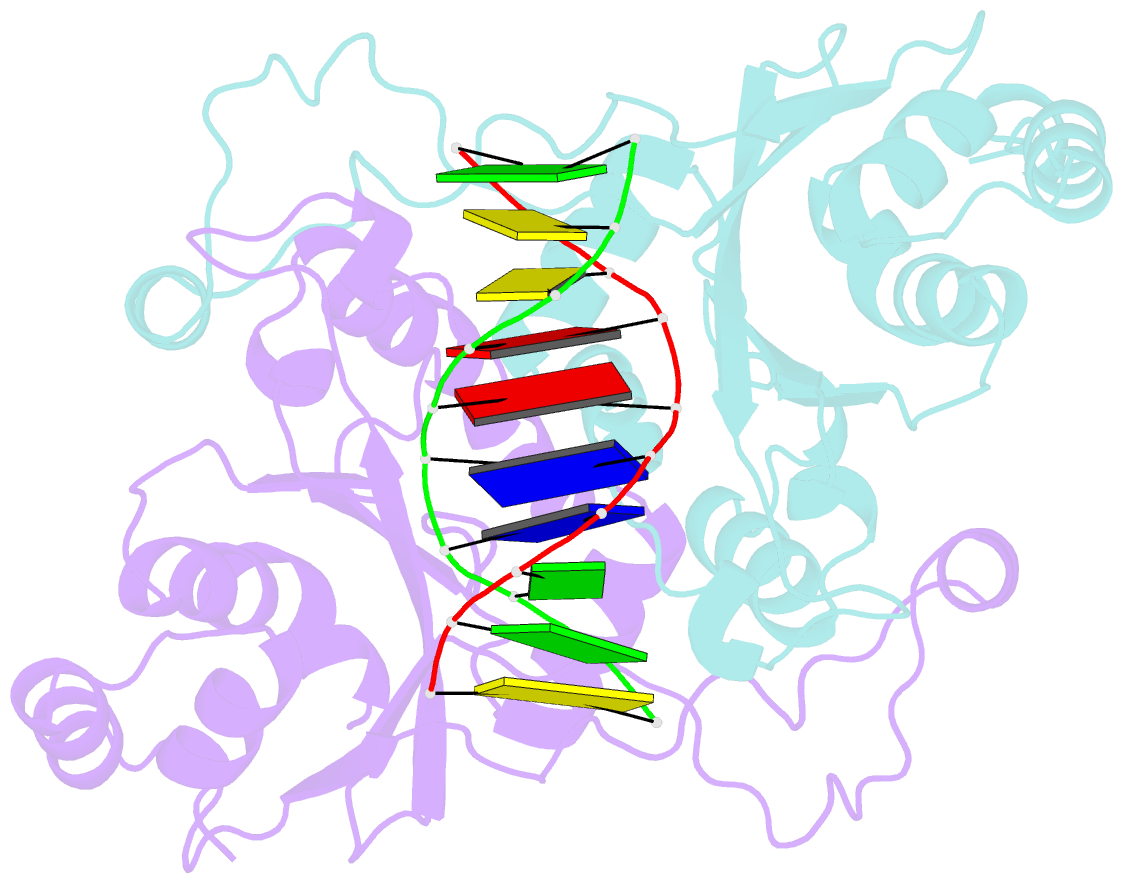
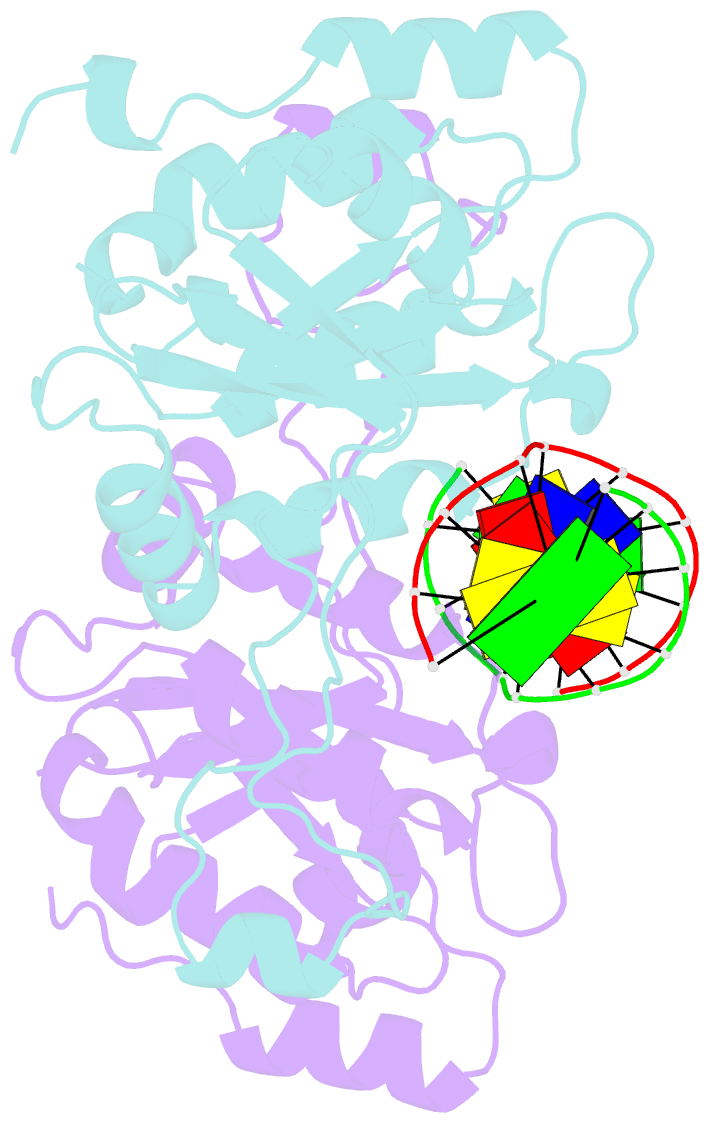
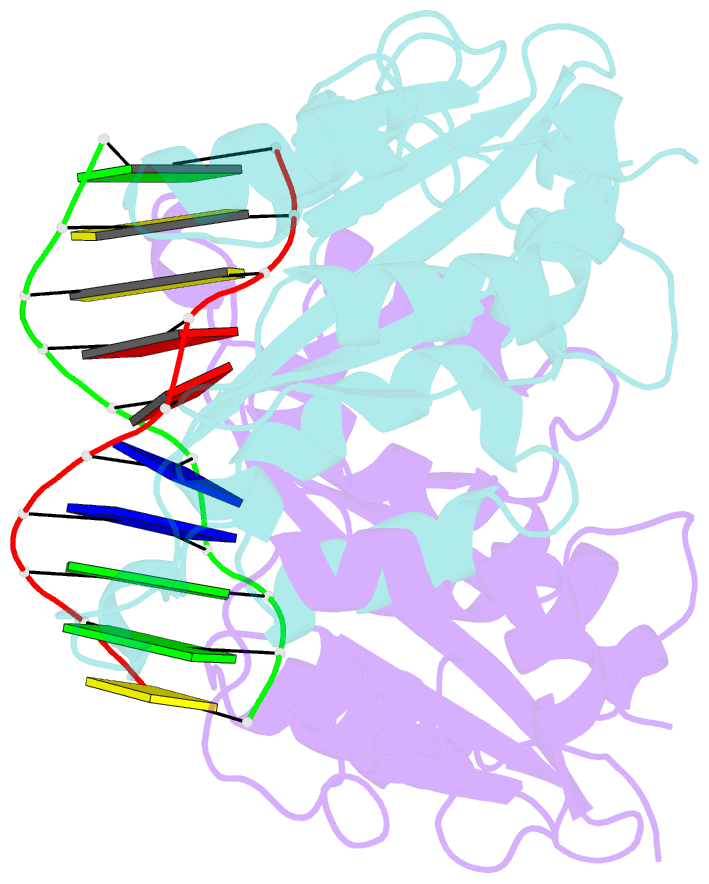
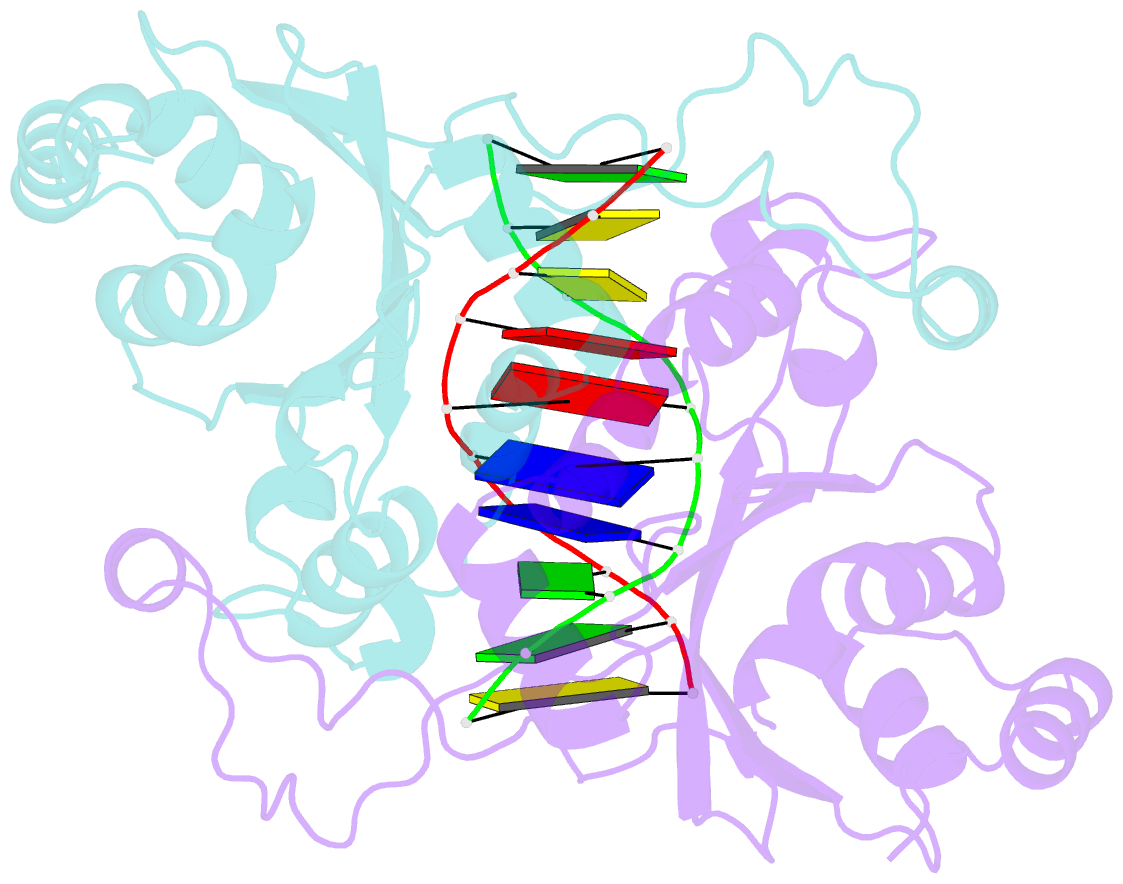
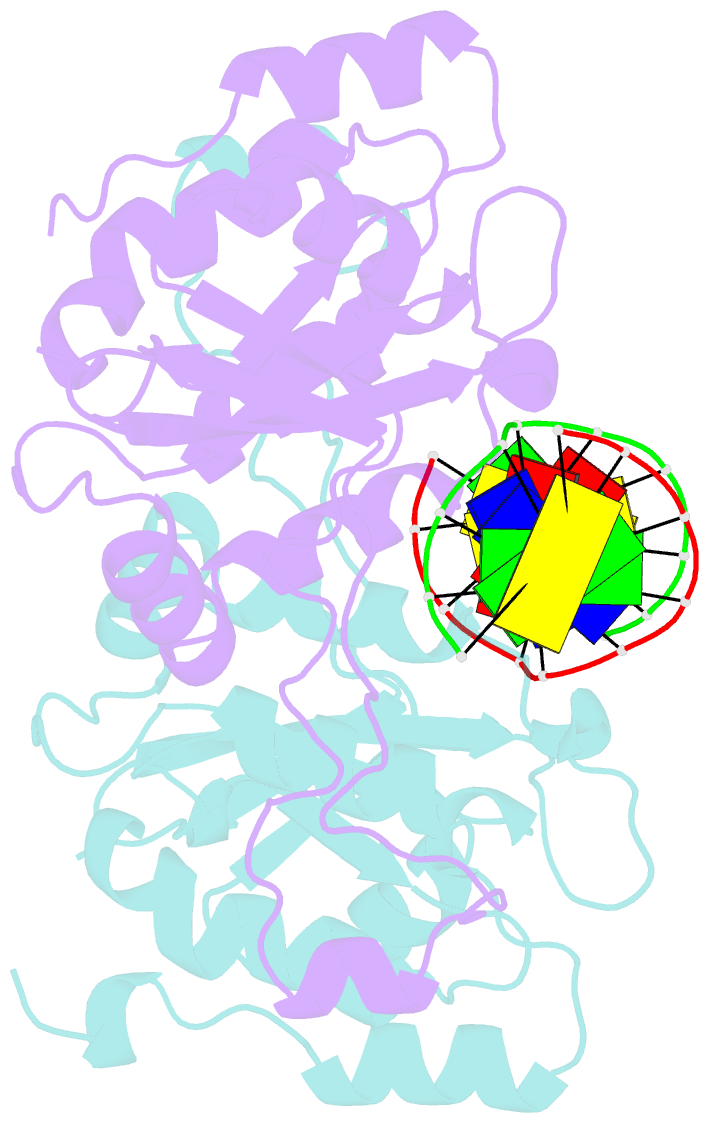
- Crystal structure of muni restriction endonuclease in complex with cognate DNA
- Reference
- Deibert M, Grazulis S, Janulaitis A, Siksnys V, Huber R (1999): "Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 A resolution." EMBO J., 18, 5805-5816. doi: 10.1093/emboj/18.21.5805.
- Abstract
- The MunI restriction enzyme recognizes the palindromic hexanucleotide sequence C/AATTG (the '/' indicates the cleavage site). The crystal structure of its active site mutant D83A bound to cognate DNA has been determined at 1.7 A resolution. Base-specific contacts between MunI and DNA occur exclusively in the major groove. While DNA-binding sites of most other restriction enzymes are comprised of discontinuous sequence segments, MunI combines all residues involved in the base-specific contacts within one short stretch (residues R115-R121) located at the N-terminal region of the 3(10)4 helix. The outer CG base pair of the recognition sequence is recognized solely by R115 through hydrogen bonds made by backbone and side chain atoms to both bases. The mechanism of recognition of the central AATT nucleotides by MunI is similar to that of EcoRI, which recognizes the G/AATTC sequence. The local conformation of AATT deviates from the typical B-DNA form and is remarkably similar to EcoRI-DNA. It appears to be essential for specific hydrogen bonding and recognition by MunI and EcoRI.