Summary information and primary citation
- PDB-id
- 1d1u; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.3 Å)
- Summary
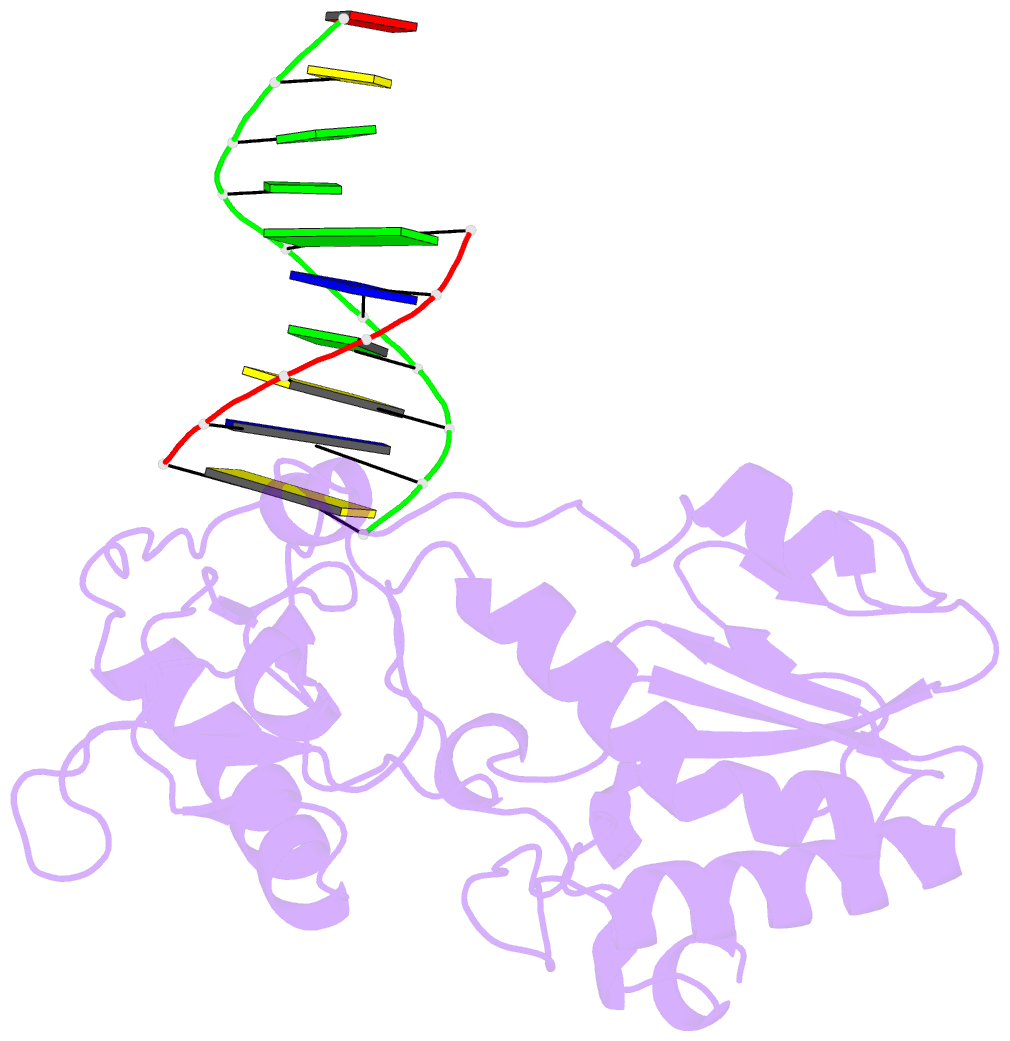
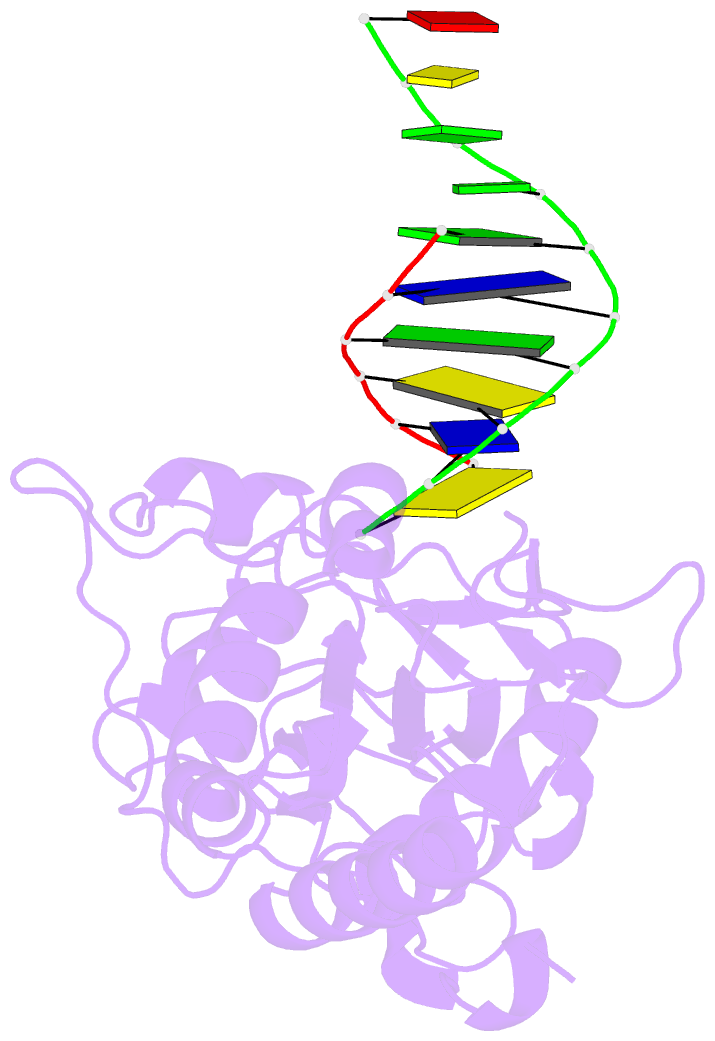
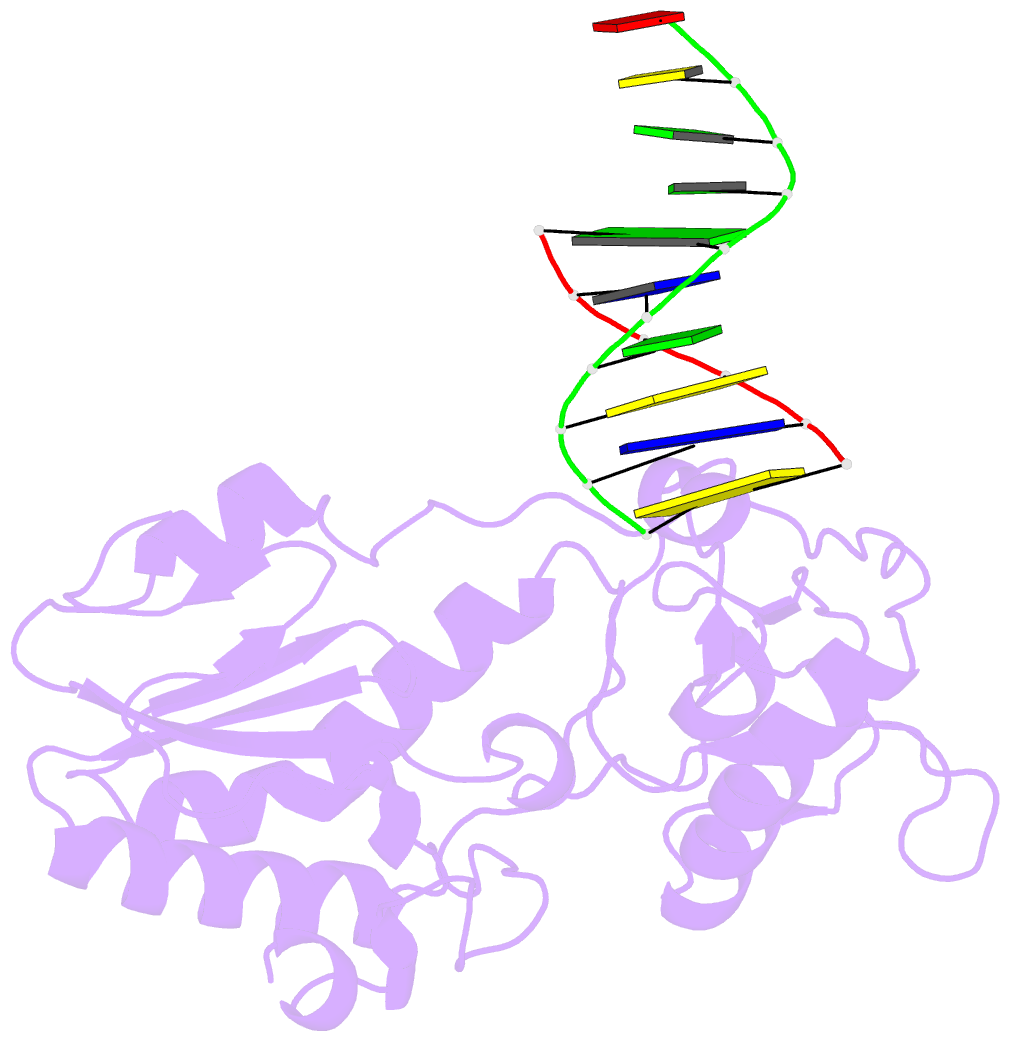
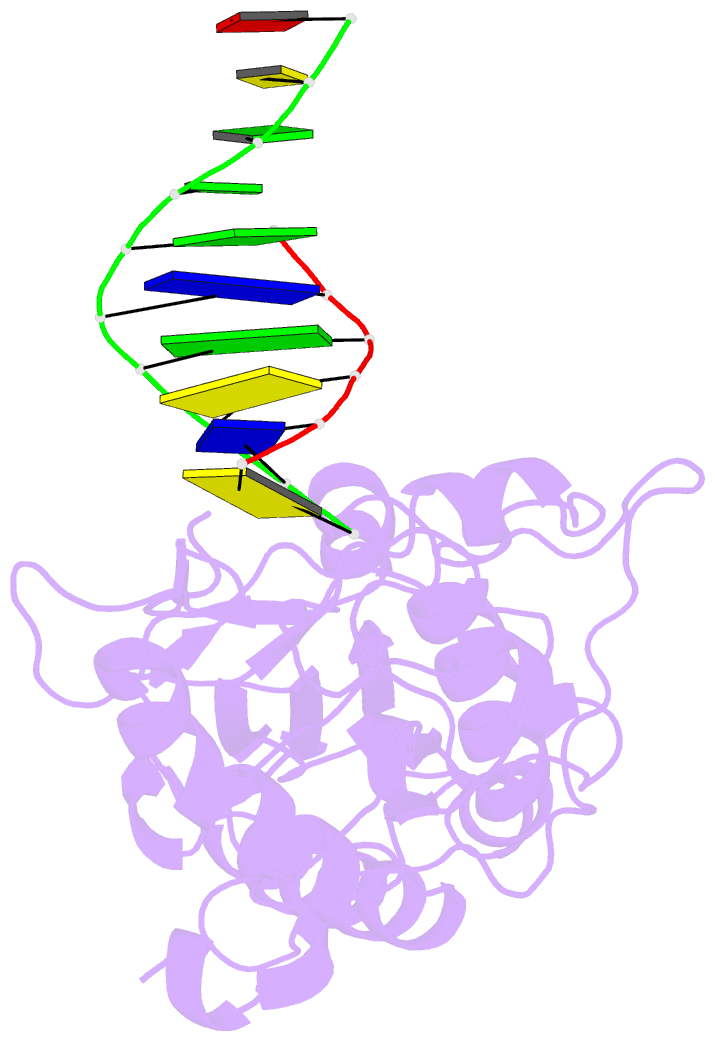
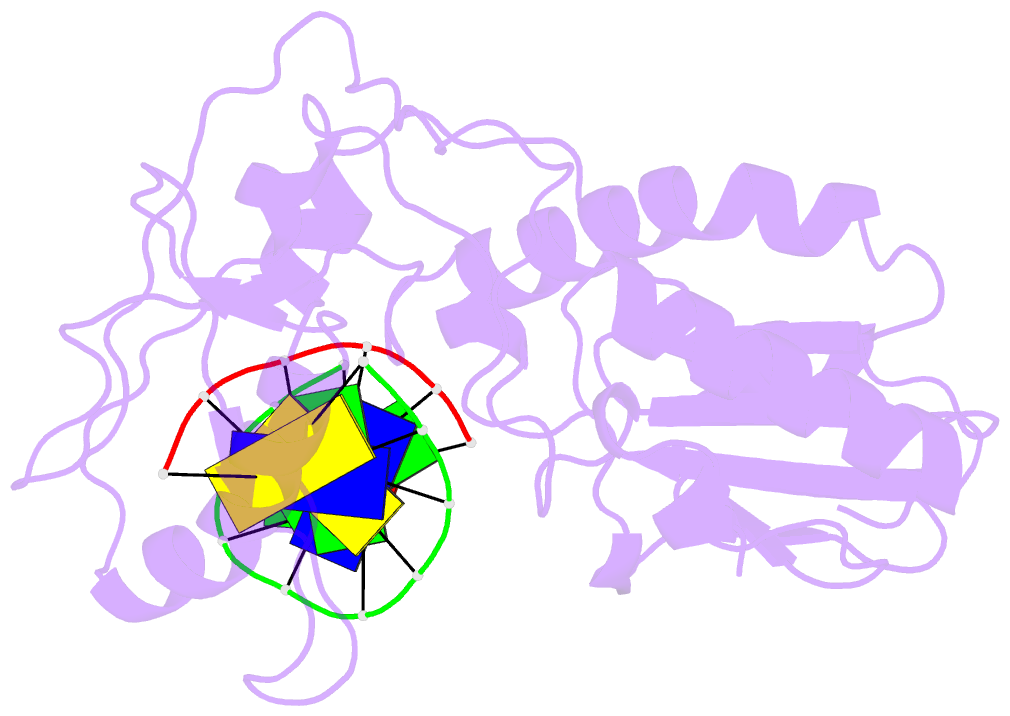
- Use of an n-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing g-a mispairs
- Reference
- Cote ML, Yohannan SJ, Georgiadis MM (2000): "Use of an N-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs." Acta Crystallogr.,Sect.D, 56, 1120-1131. doi: 10.1107/S0907444900008246.
- Abstract
- Complexation with the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase offers a novel method of obtaining crystal structures of nucleic acid duplexes, which can be phased by molecular replacement. This method is somewhat similar to the method of using a monoclonal antibody Fab fragment complexed to the molecule of interest in order to obtain crystals suitable for X-ray crystallographic analysis. Here a novel DNA structure including two G-A mispairs in a pseudo-hexadecamer determined at 2.3 A resolution in a complex with the N-terminal fragment is reported. This structure has an asymmetric unit consisting of the protein molecule bound to the blunt end of a DNA 6/10-mer, which is composed of a six-base strand (5'-CTCGTG-3') and a ten-base strand (3'-GAGCACGGCA-5'). The 6/10-mer is thus composed of a six-base-pair duplex with a four-base single-stranded overhang. In the crystal structure, the bases of the overhang are reciprocally paired (symmetry element -x - 1, -y, z), yielding a doubly nicked pseudo-hexadecamer primarily B-form DNA molecule, which has some interesting A-like structural features. The pairing between the single strands results in two standard (G-C) Watson-Crick pairs and two G-A mispairs. The structural DNA model can accommodate either a standard syn or a standard anti conformation for the 5'-terminal adenine of the ten-base strand of the DNA based on analysis of simulated-annealing omit maps. Although the DNA model here includes nicks in the phosphodiester backbone, modeling of an intact phosphodiester backbone results in a very similar DNA model and indicates that the structure is biologically relevant.