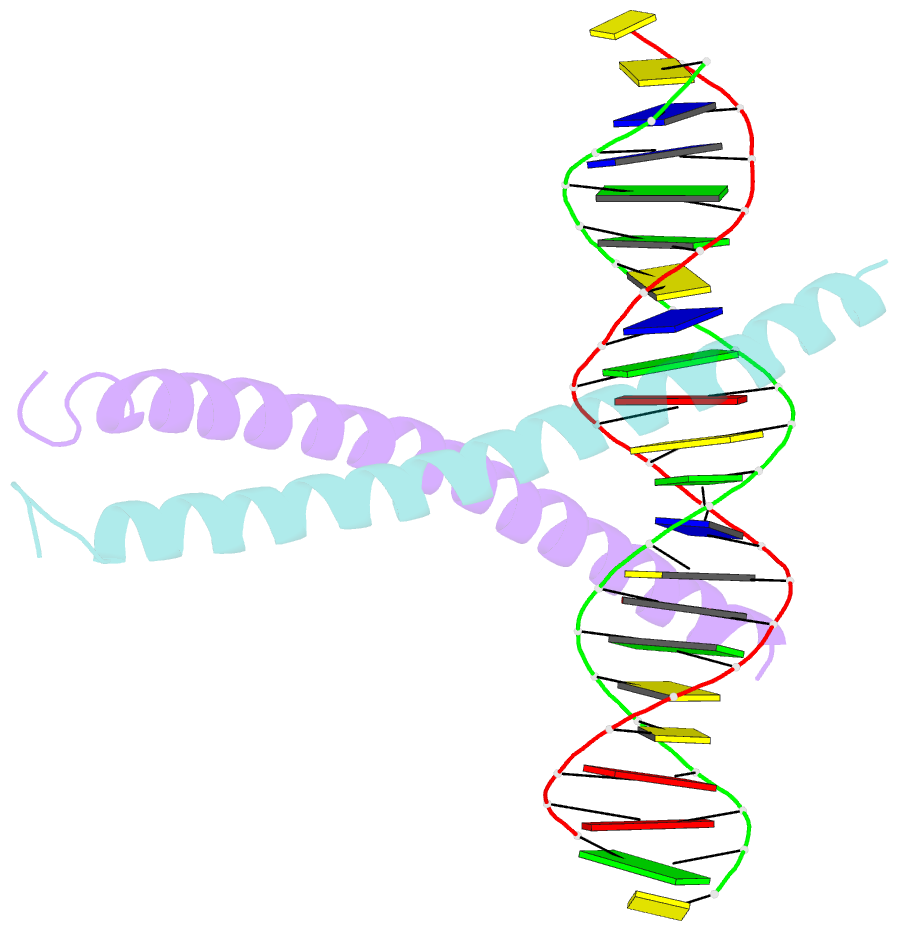
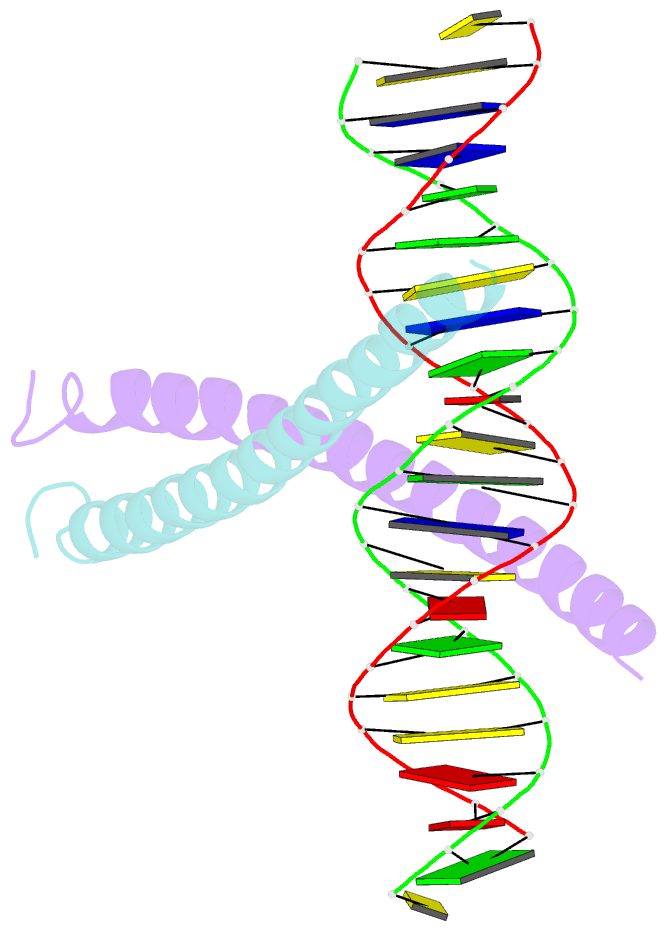
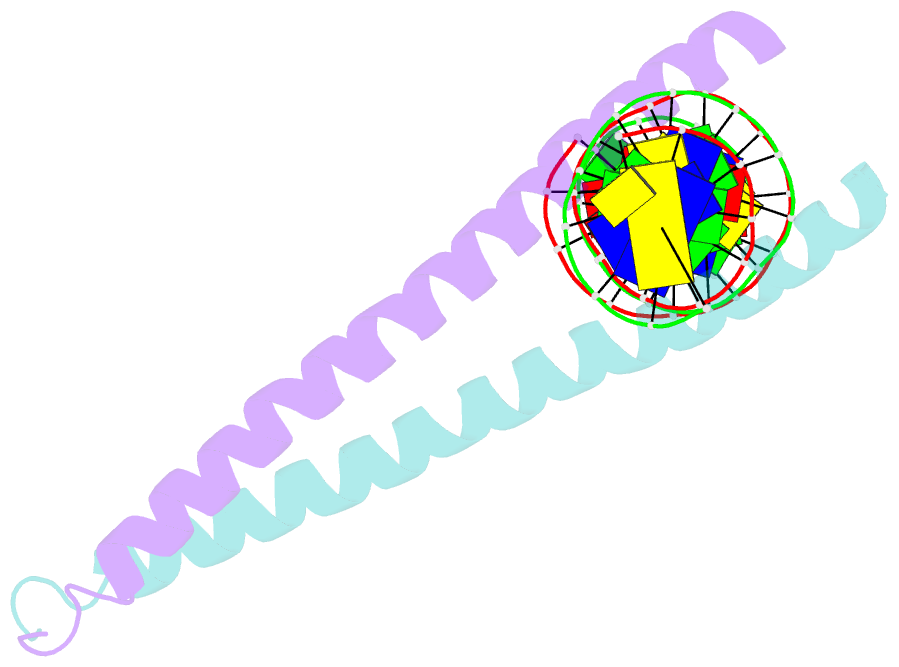
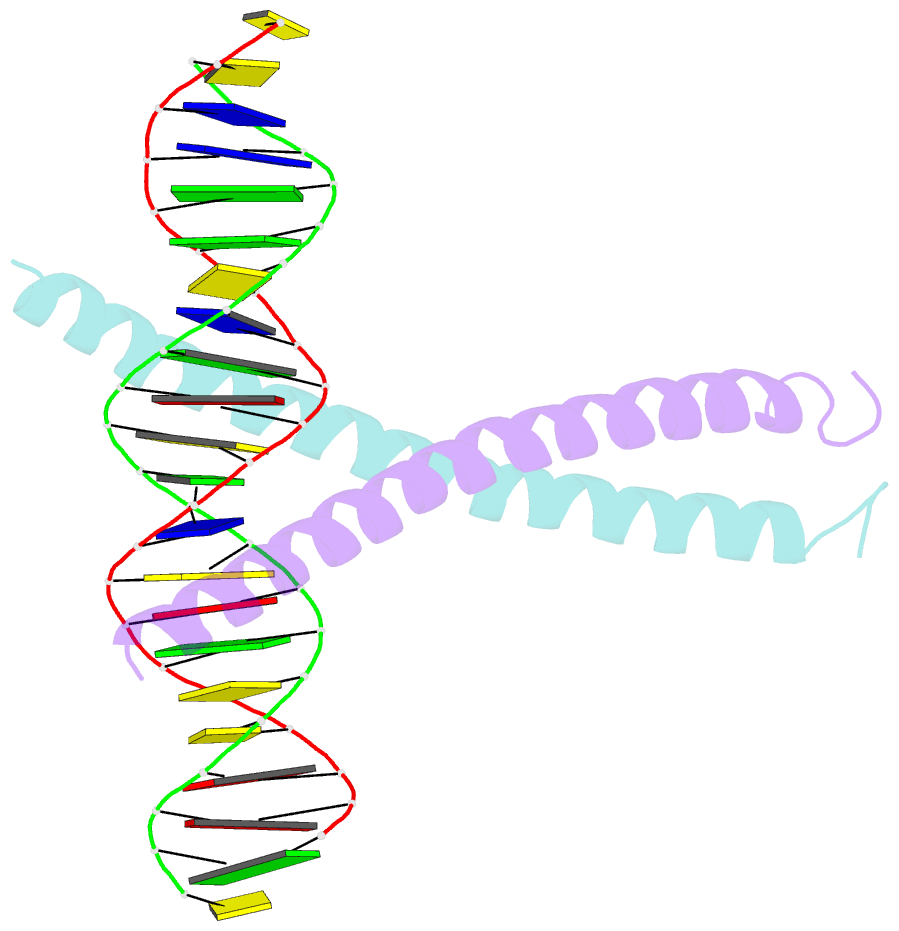
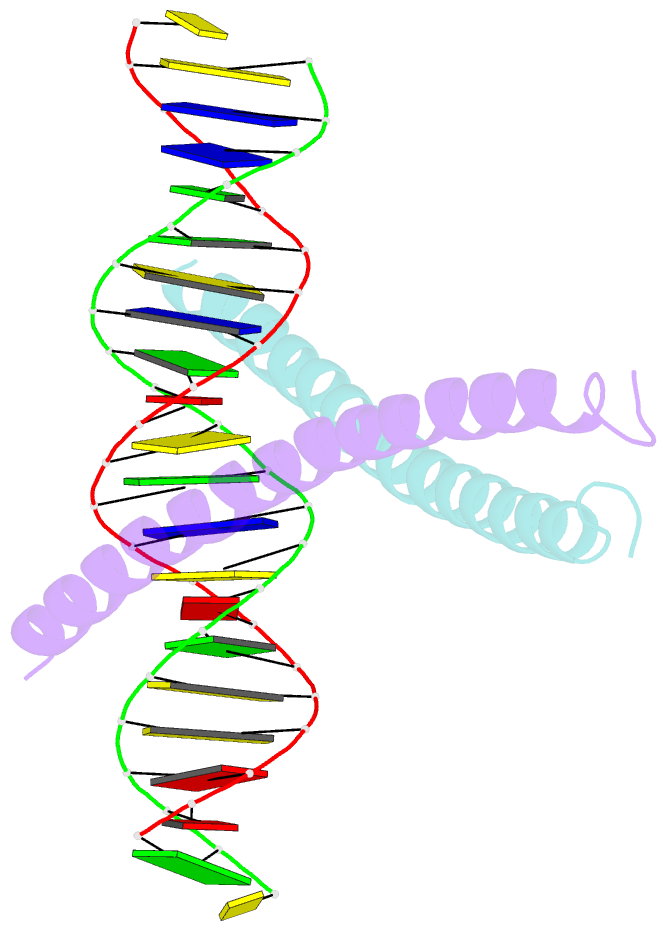
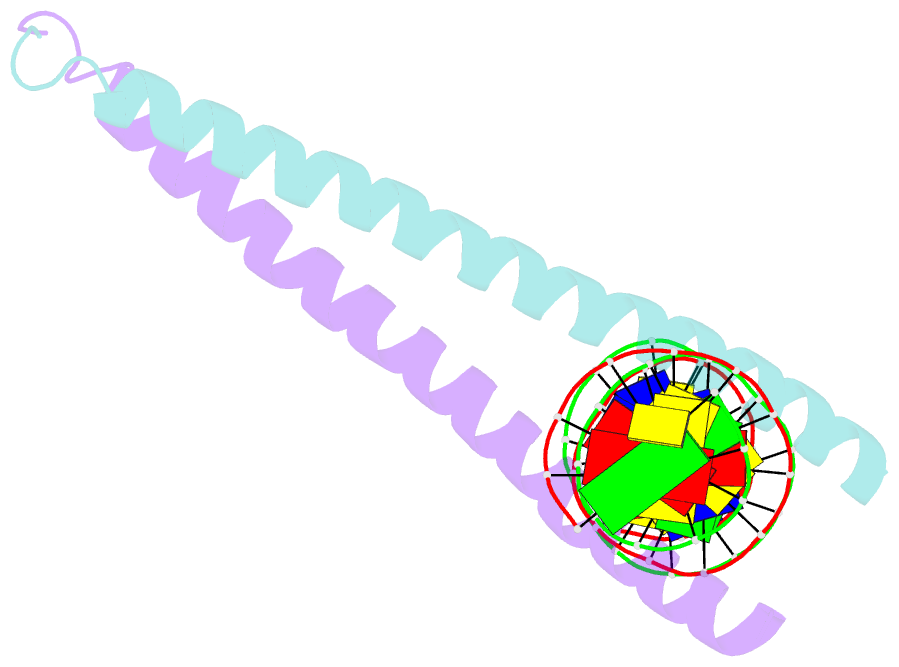
Summary information and primary citation
- PDB-id
- 1dh3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.0 Å)
- Summary
- Crystal structure of a creb bzip-cre complex reveals the basis for creb faimly selective dimerization and DNA binding
- Reference
- Schumacher MA, Goodman RH, Brennan RG (2000): "The structure of a CREB bZIP.somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding." J.Biol.Chem., 275, 35242-35247. doi: 10.1074/jbc.M007293200.
- Abstract
- The cAMP responsive element-binding protein (CREB) is central to second messenger regulated transcription. To elucidate the structural mechanisms of DNA binding and selective dimerization of CREB, we determined to 3.0 A resolution, the structure of the CREB bZIP (residues 283-341) bound to a 21-base pair deoxynucleotide that encompasses the canonical 8-base pair somatostatin cAMP response element (SSCRE). The CREB dimer is stabilized in part by ionic interactions from Arg(314) to Glu(319') and Glu(328) to Lys(333') as well as a hydrogen bond network that links the carboxamide side chains of Gln(322')-Asn(321)-Asn(321')-Gln(322). Critical to family selective dimerization are intersubunit hydrogen bonds between basic region residue Tyr(307) and leucine zipper residue Glu(312), which are conserved in all CREB/CREM/ATF-1 family members. Strikingly, the structure reveals a hexahydrated Mg(2+) ion bound in the cavity between the basic region and SSCRE that makes a water-mediated DNA contact. DNA binding studies demonstrate that Mg(2+) ions enhance CREB bZIP:SSCRE binding by more than 25-fold and suggest a possible physiological role for this ion in somatostatin cAMP response element and potentially other CRE-mediated gene expression.