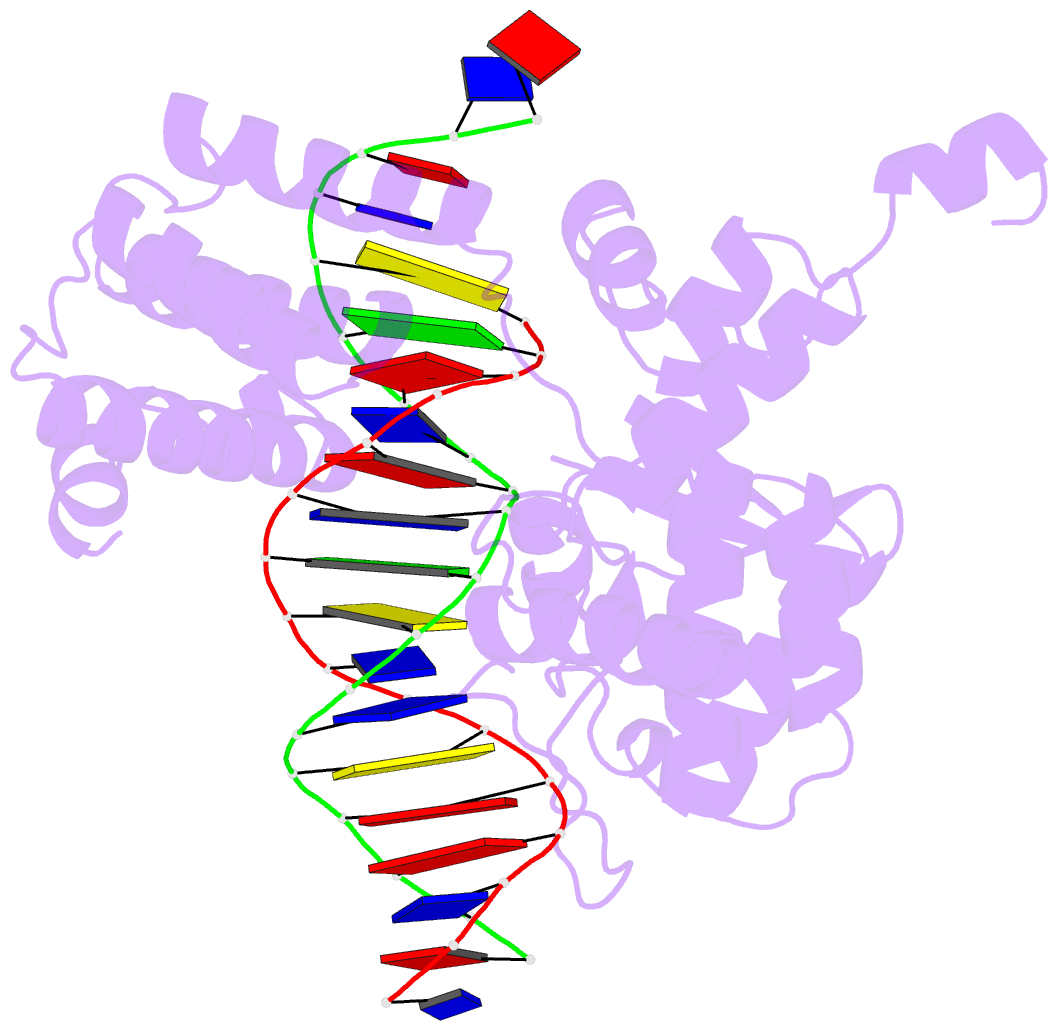
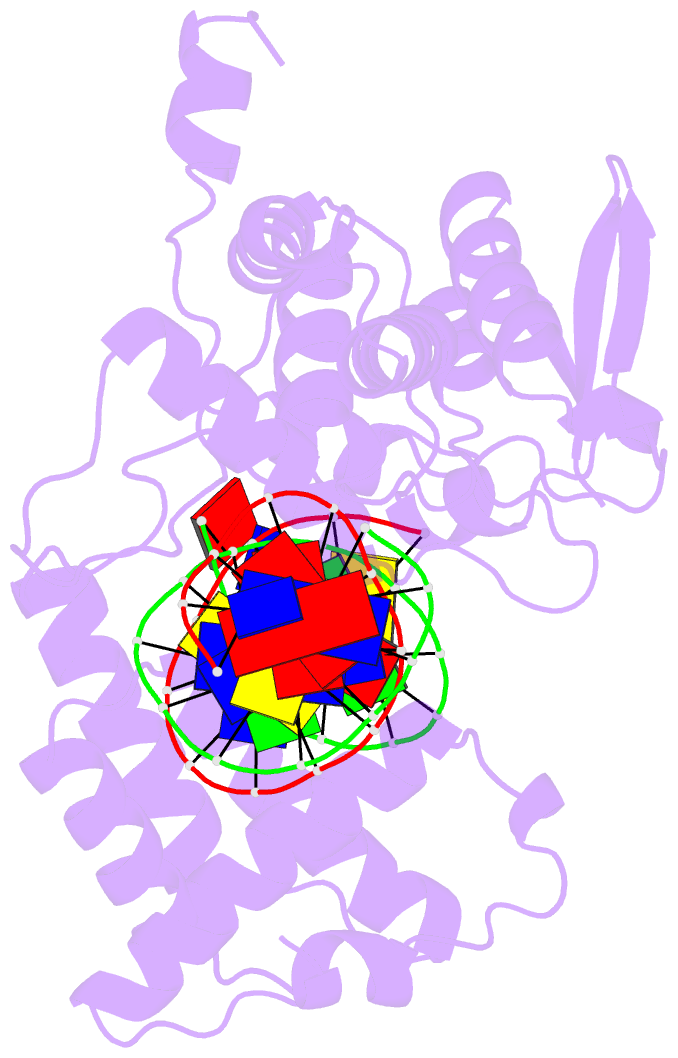
Summary information and primary citation
- PDB-id
- 1drg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase, ligase-DNA
- Method
- X-ray (2.55 Å)
- Summary
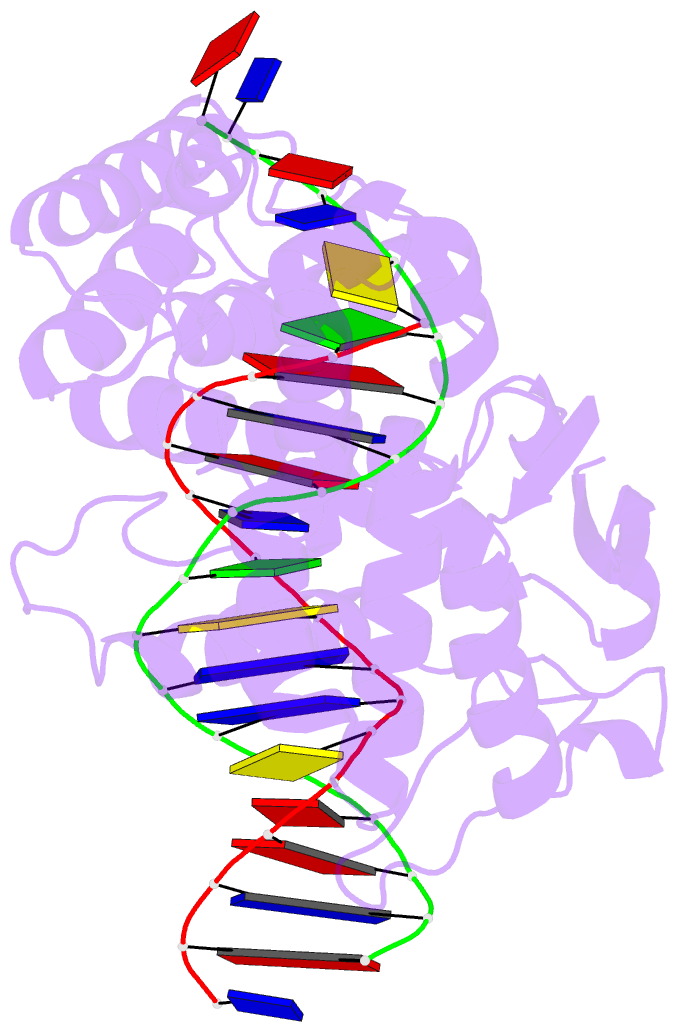
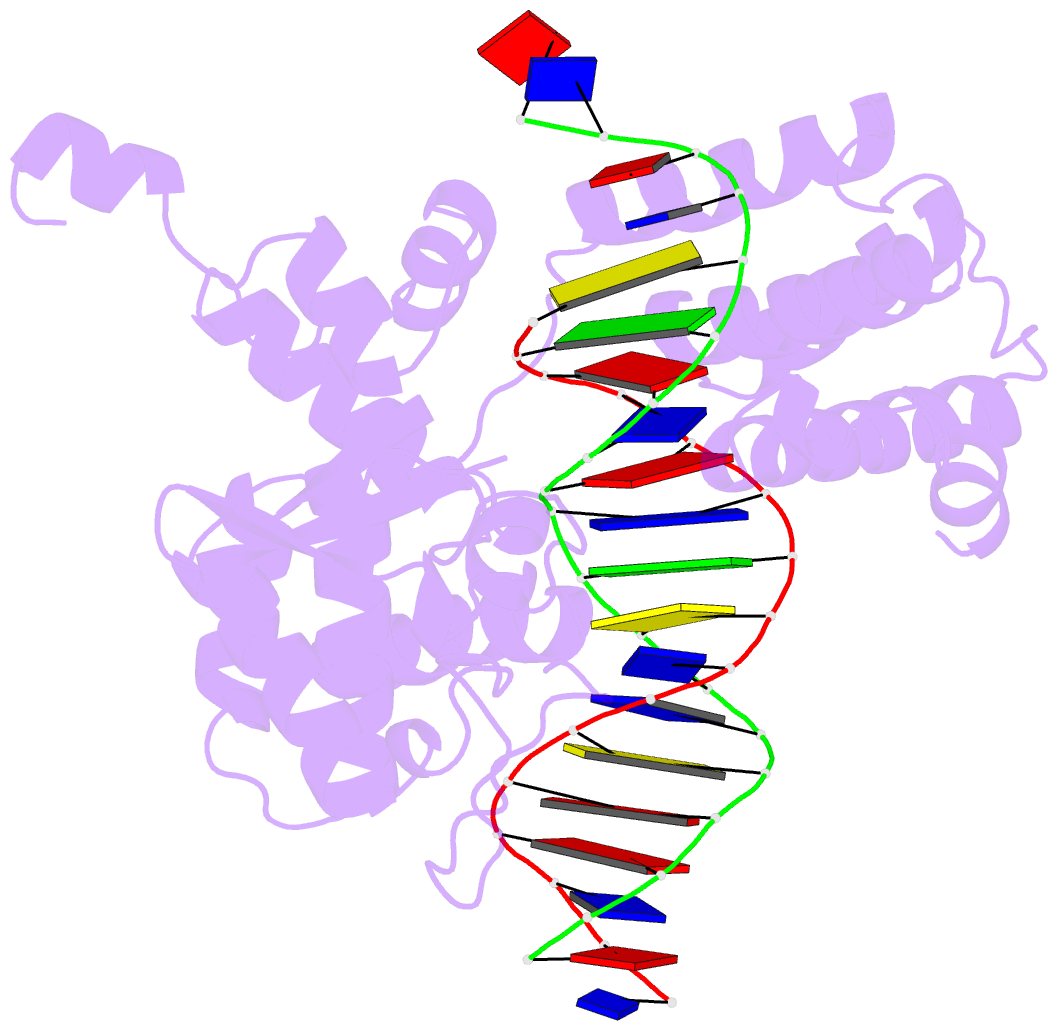
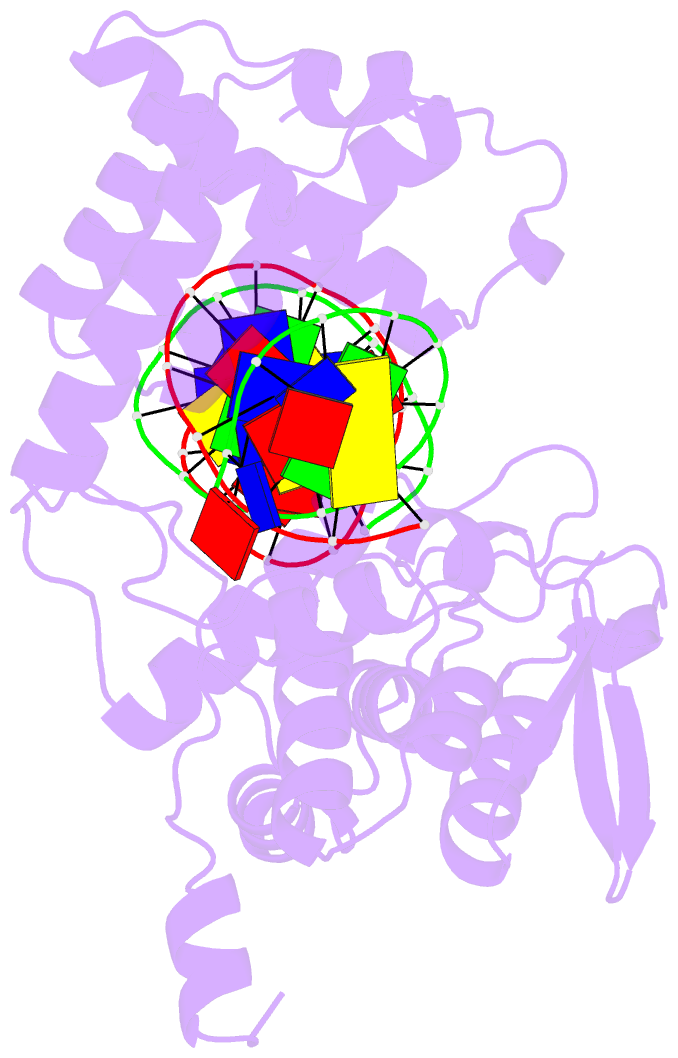
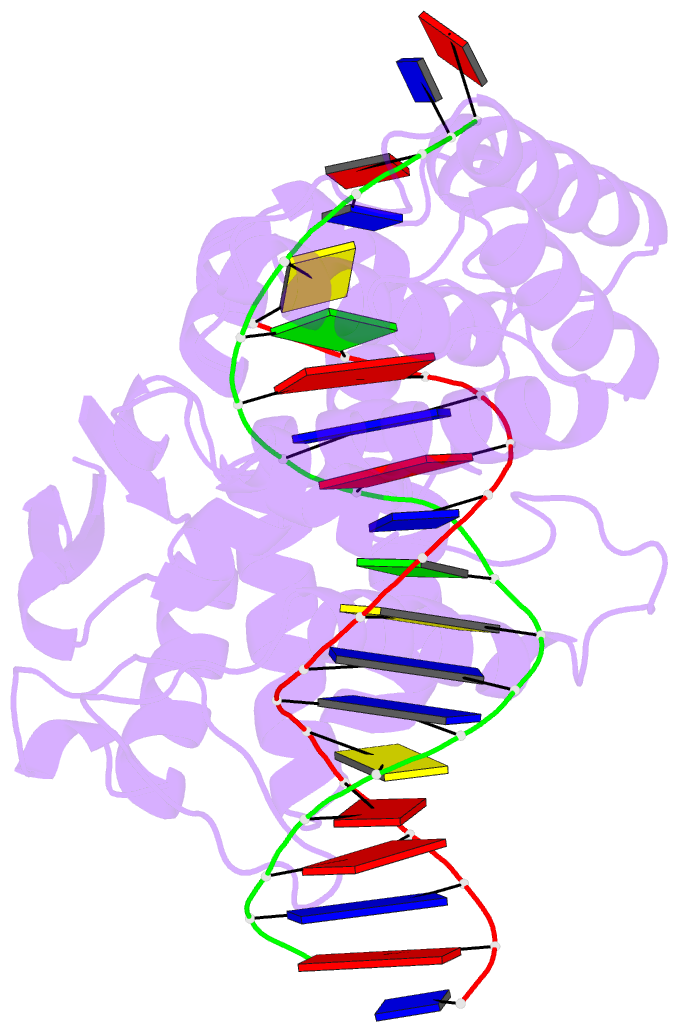
- Crystal structure of trimeric cre recombinase-lox complex
- Reference
- Woods KC, Martin SS, Chu VC, Baldwin EP (2001): "Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction." J.Mol.Biol., 313, 49-69. doi: 10.1006/jmbi.2001.5012.
- Abstract
- The crystal structure of a novel Cre-Lox synapse was solved using phases from multiple isomorphous replacement and anomalous scattering, and refined to 2.05 A resolution. In this complex, a symmetric protein trimer is bound to a Y-shaped three-way DNA junction, a marked departure from the pseudo-4-fold symmetrical tetramer associated with Cre-mediated LoxP recombination. The three-way DNA junction was accommodated by a simple kink without significant distortion of the adjoining DNA duplexes. Although the mean angle between DNA arms in the Y and X structures was similar, adjacent Cre trimer subunits rotated 29 degrees relative to those in the tetramers. This rotation was accommodated at the protein-protein and DNA-DNA interfaces by interactions that are "quasi-equivalent" to those in the tetramer, analogous to packing differences of chemically identical viral subunits at non-equivalent positions in icosahedral capsids. This structural quasi-equivalence extends to function as Cre can bind to, cleave and perform strand transfer with a three-way Lox substrate. The structure explains the dual recognition of three and four-way junctions by site-specific recombinases as being due to shared structural features between the differently branched substrates and plasticity of the protein-protein interfaces. To our knowledge, this is the first direct demonstration of quasi-equivalence in both the assembly and function of an oligomeric enzyme.