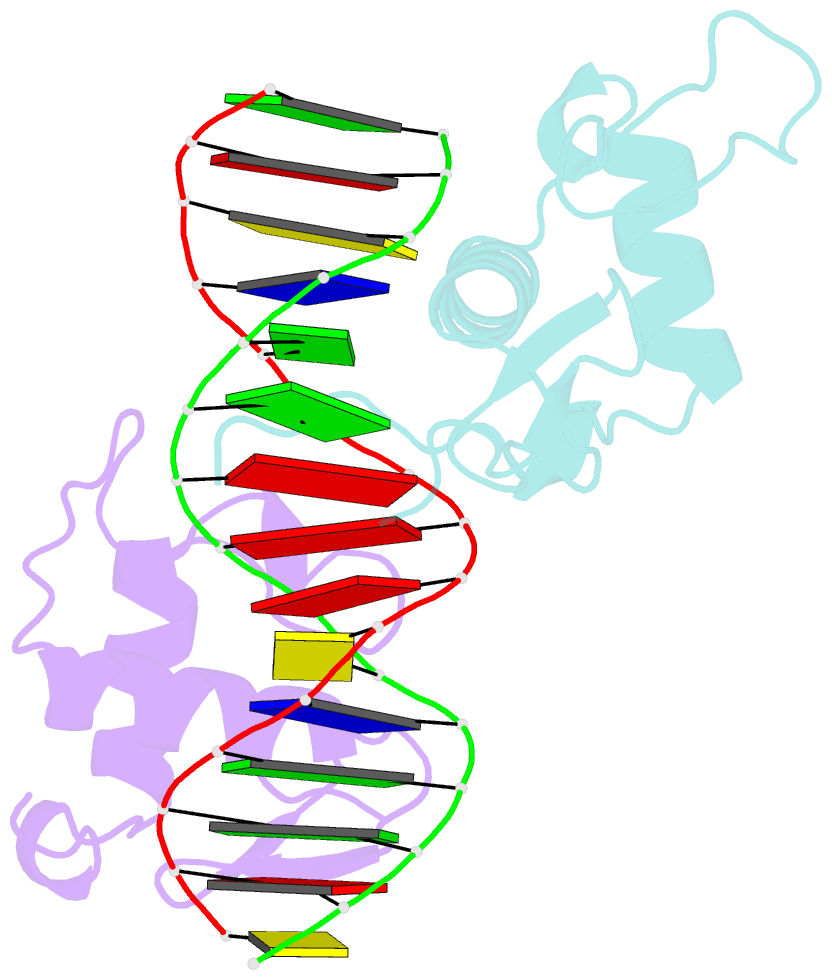
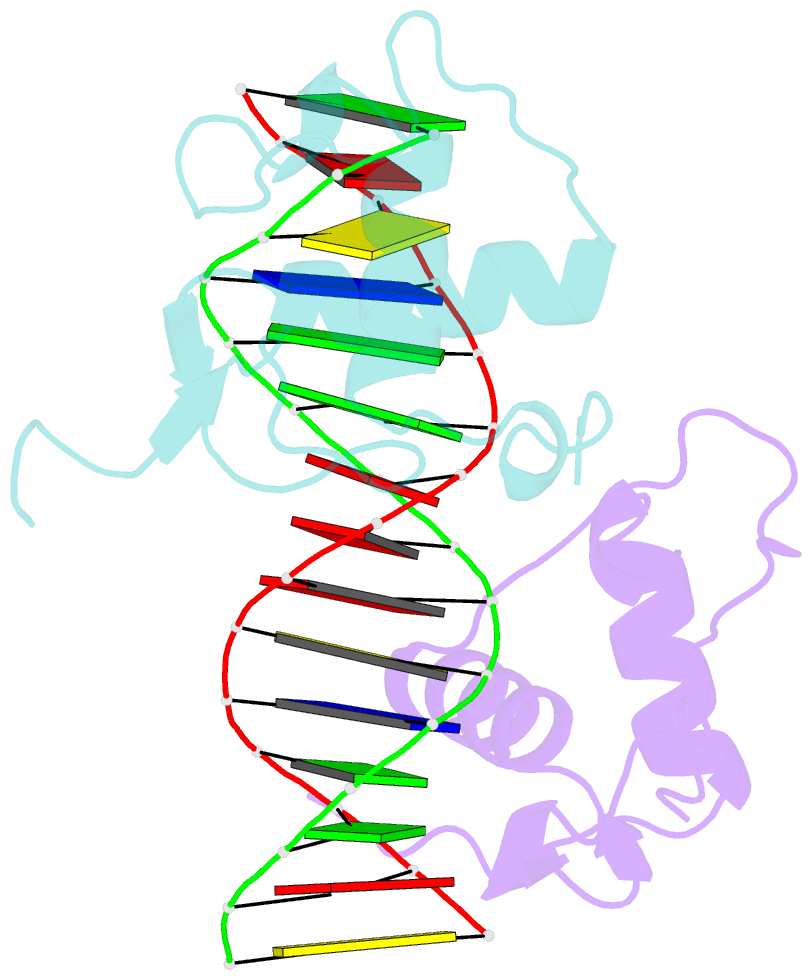
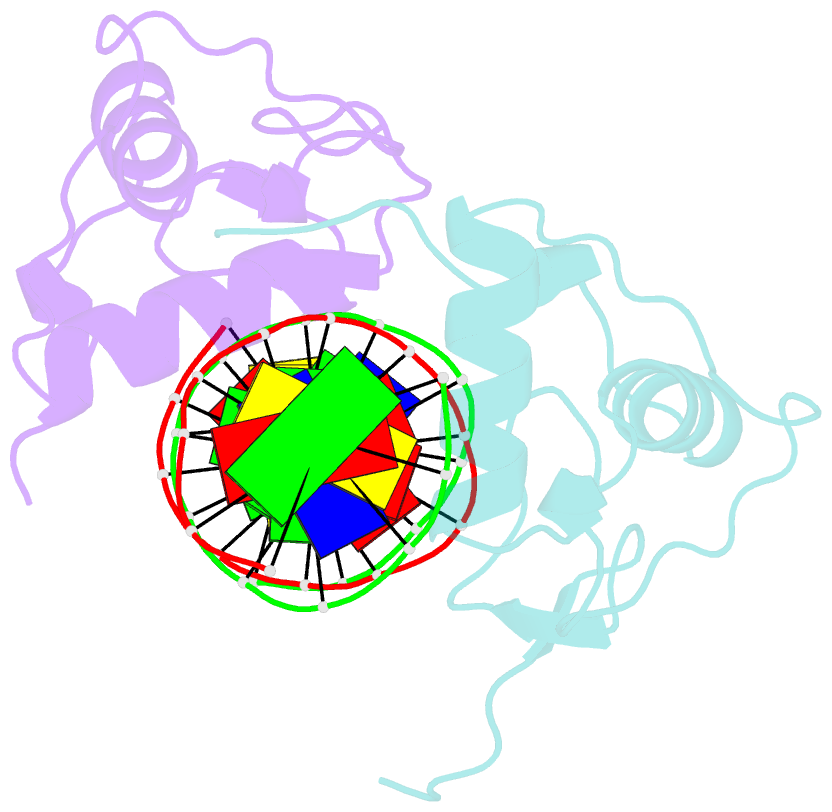
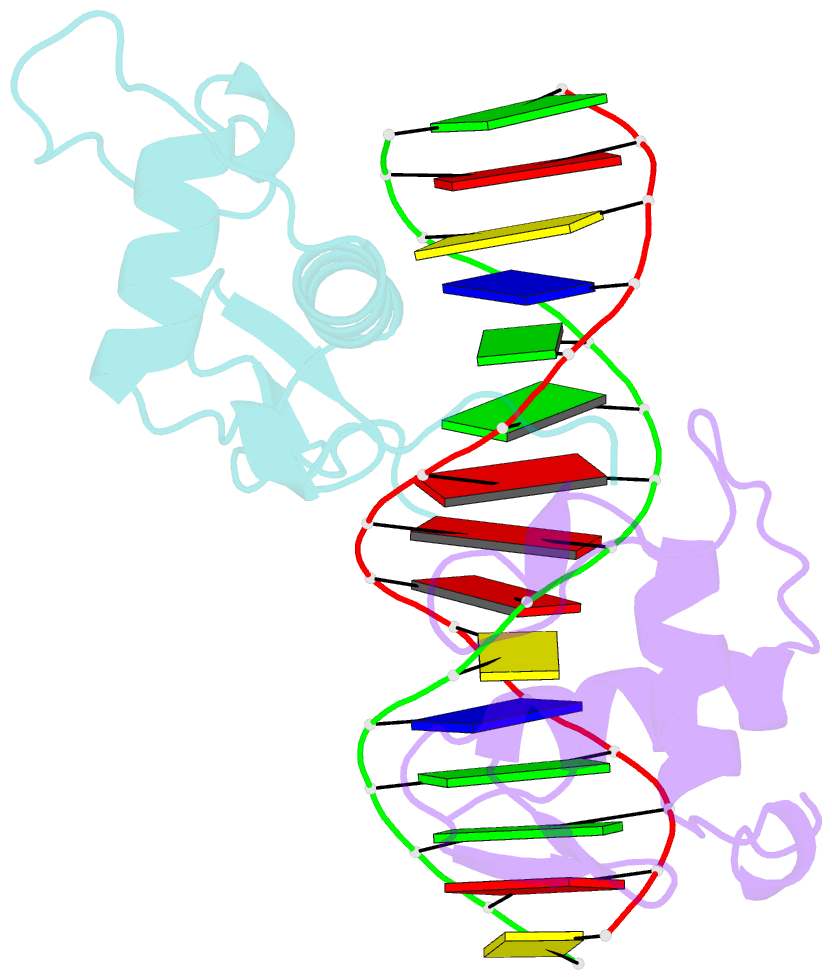
Summary information and primary citation
- PDB-id
- 1dsz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (1.7 Å)
- Summary
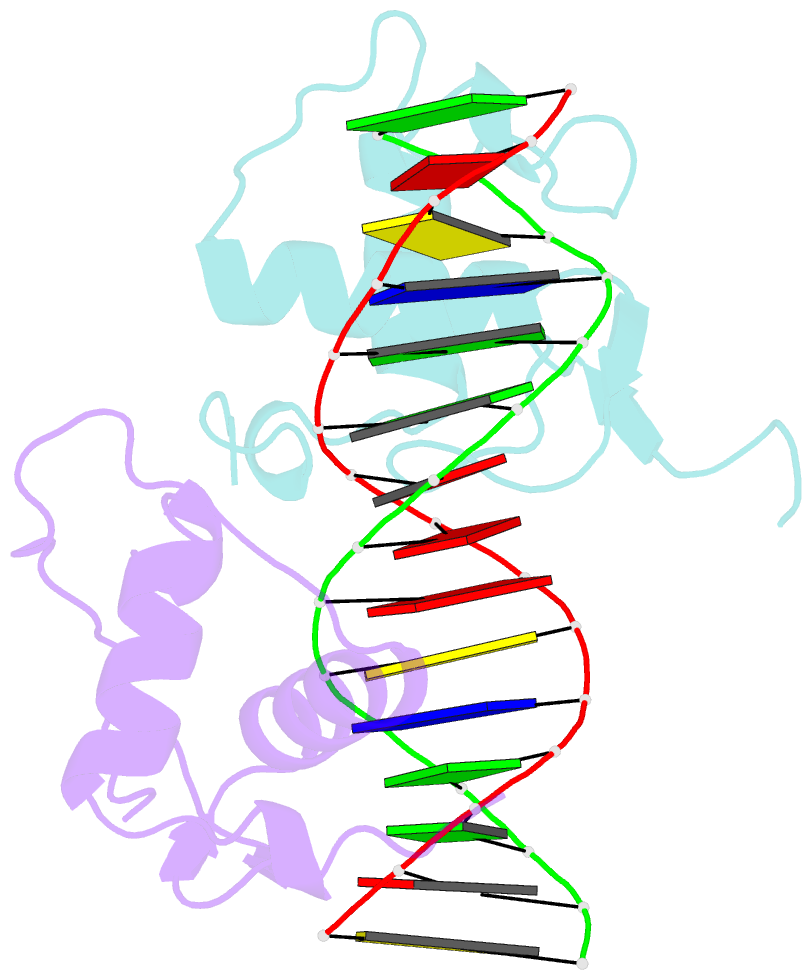
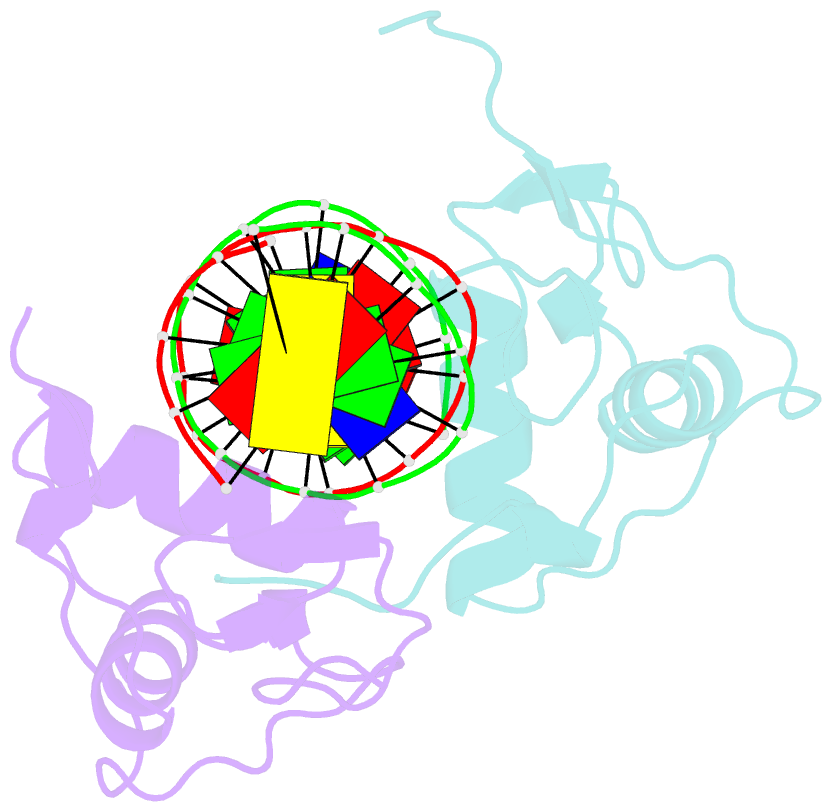
- Structure of the rxr-rar DNA-binding domain heterodimer in complex with the retinoic acid response element dr1
- Reference
- Rastinejad F, Wagner T, Zhao Q, Khorasanizadeh S (2000): "Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1." EMBO J., 19, 1045-1054. doi: 10.1093/emboj/19.5.1045.
- Abstract
- The 9-cis retinoic acid receptor (retinoid X receptor, RXR) forms heterodimers with the all-trans retinoic acid receptor (RAR) and other nuclear receptors on DNA regulatory sites composed of tandem binding elements. We describe the 1.70 A resolution structure of the ternary complex of RXR and RAR DNA-binding regions in complex with the retinoic acid response element DR1. The receptors recognize identical half-sites through extensive base-specific contacts; however, RXR binds exclusively to the 3' site to form an asymmetric complex with the reverse polarity of other RXR heterodimers. The subunits associate in a strictly DNA-dependent manner using the T-box of RXR and the Zn-II region of RAR, both of which are reshaped in forming the complex. The protein-DNA contacts, the dimerization interface and the DNA curvature in the RXR-RAR complex are distinct from those of the RXR homodimer, which also binds DR1. Together, these structures illustrate how the nuclear receptor superfamily exploits conformational flexibility and locally induced structures to generate combinatorial transcription factors.