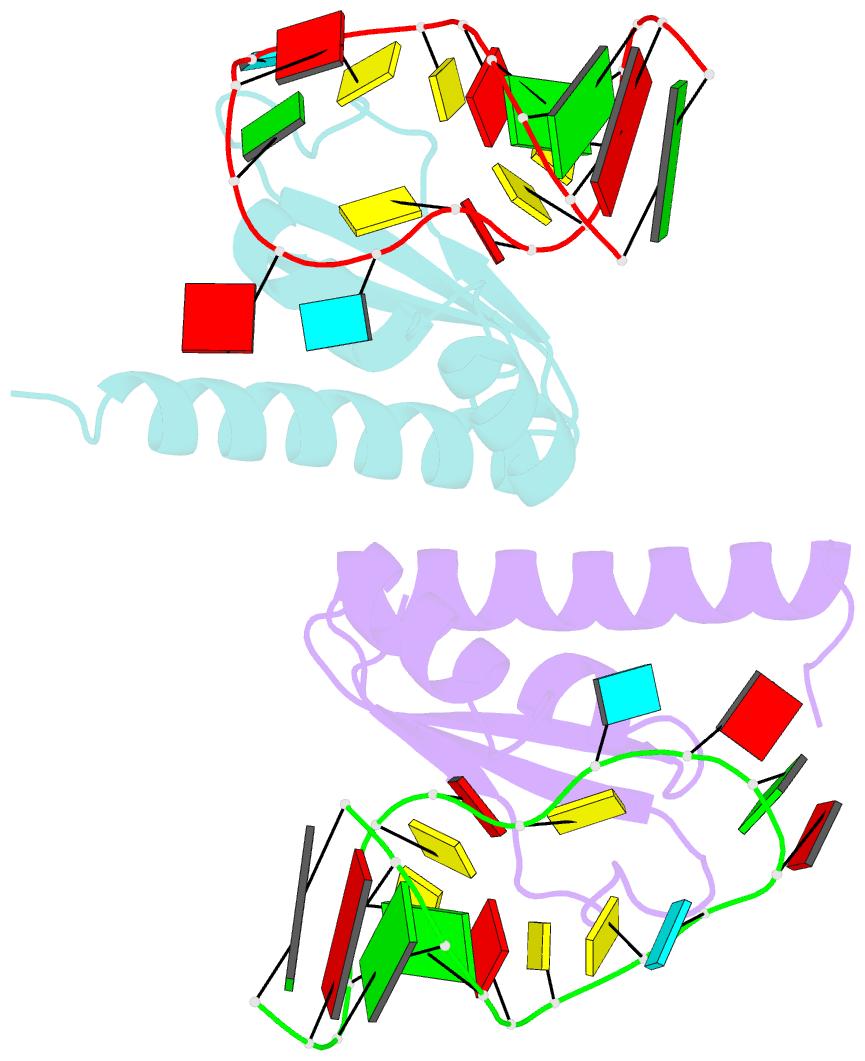
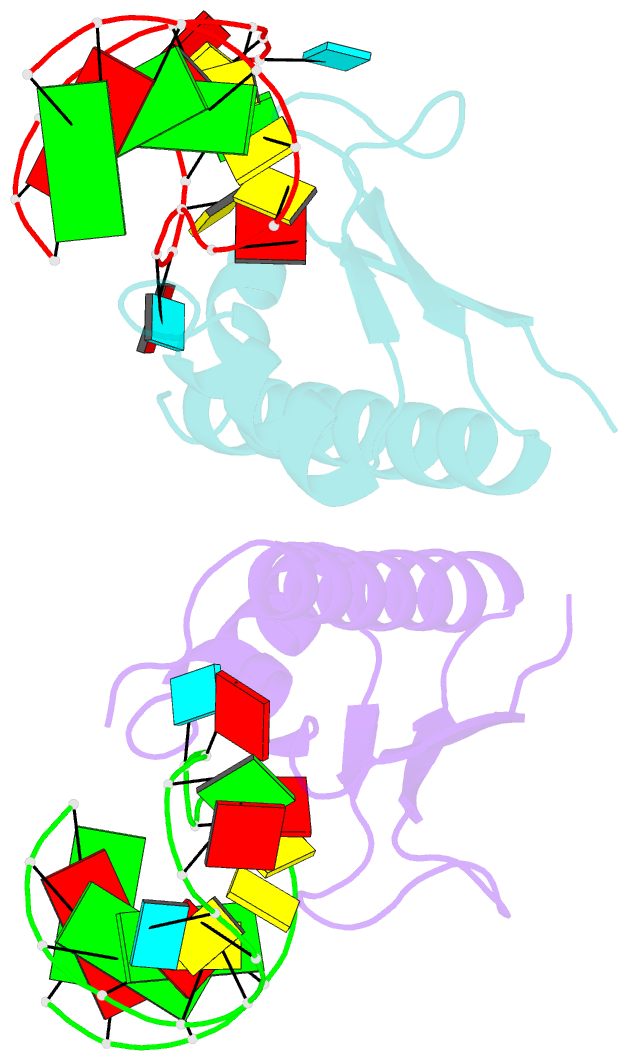
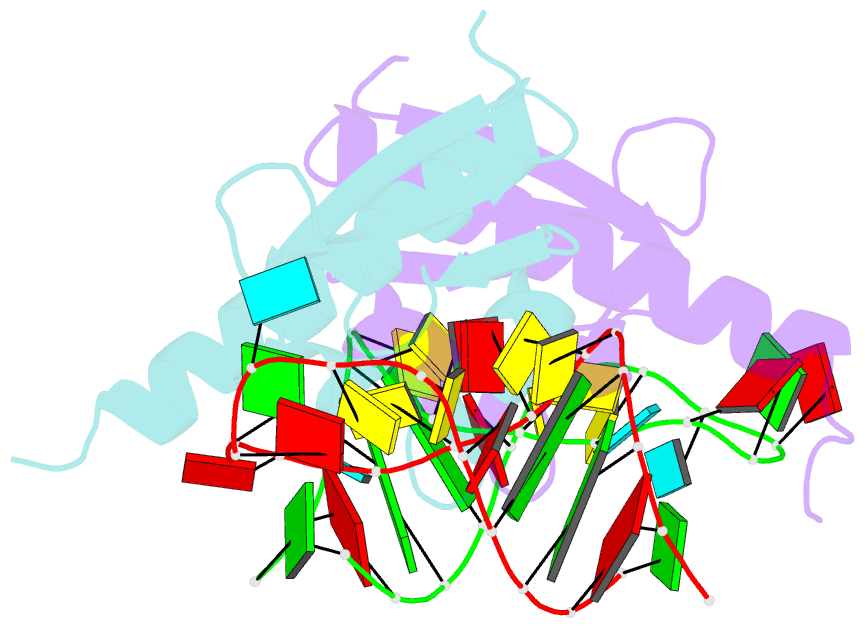
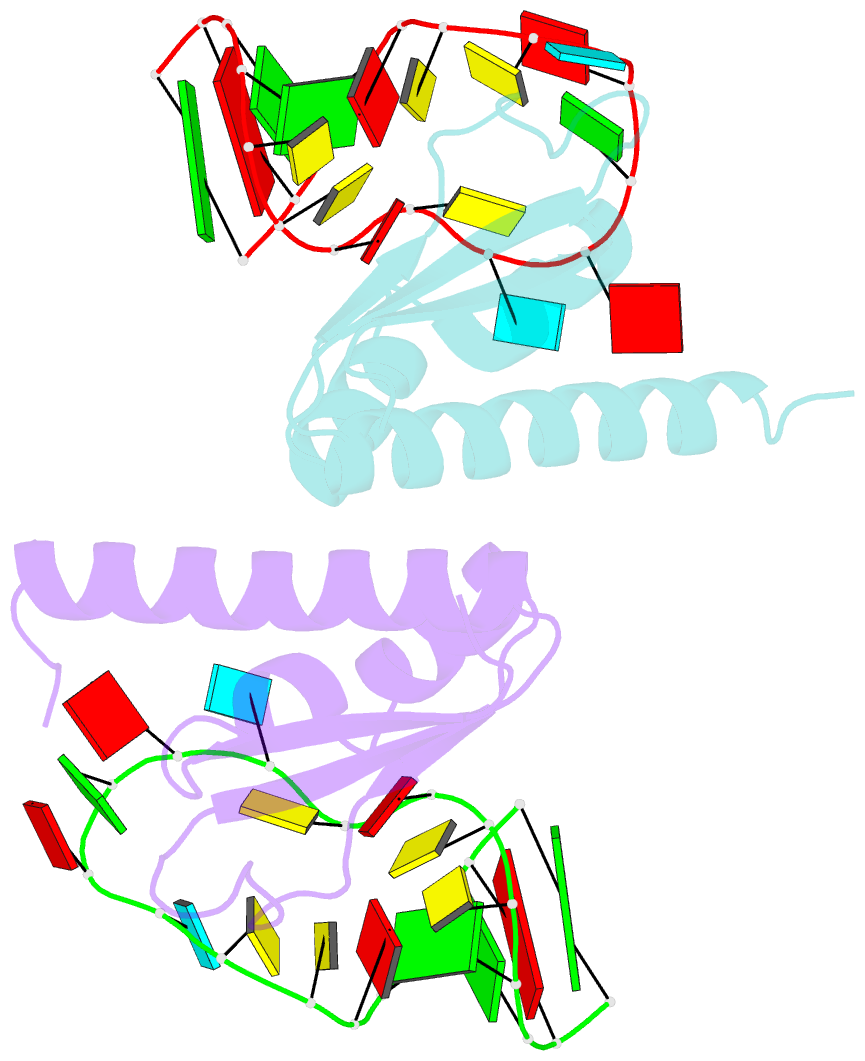
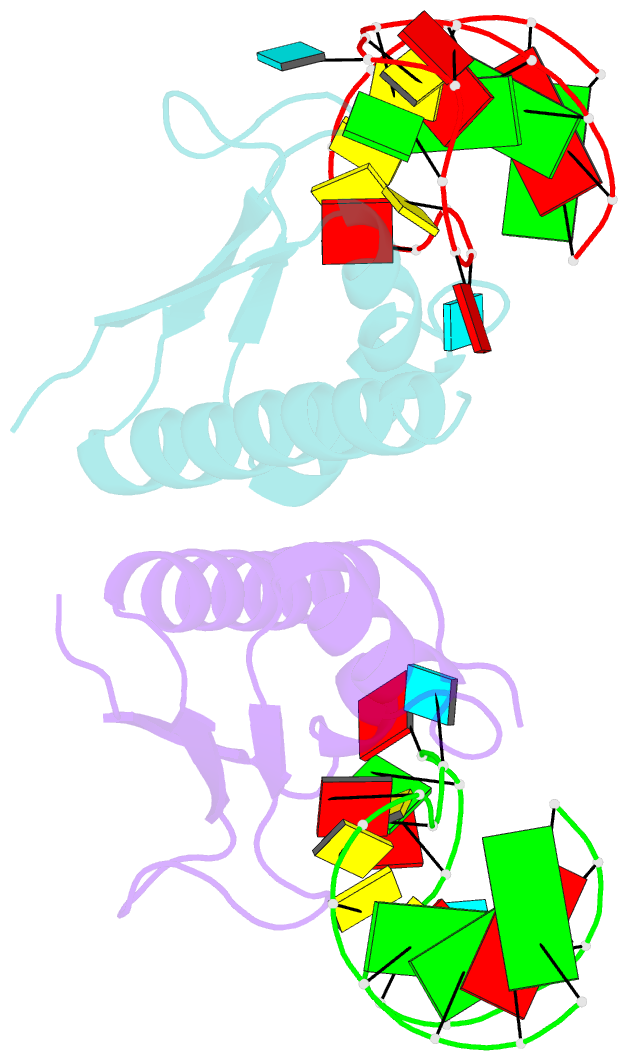
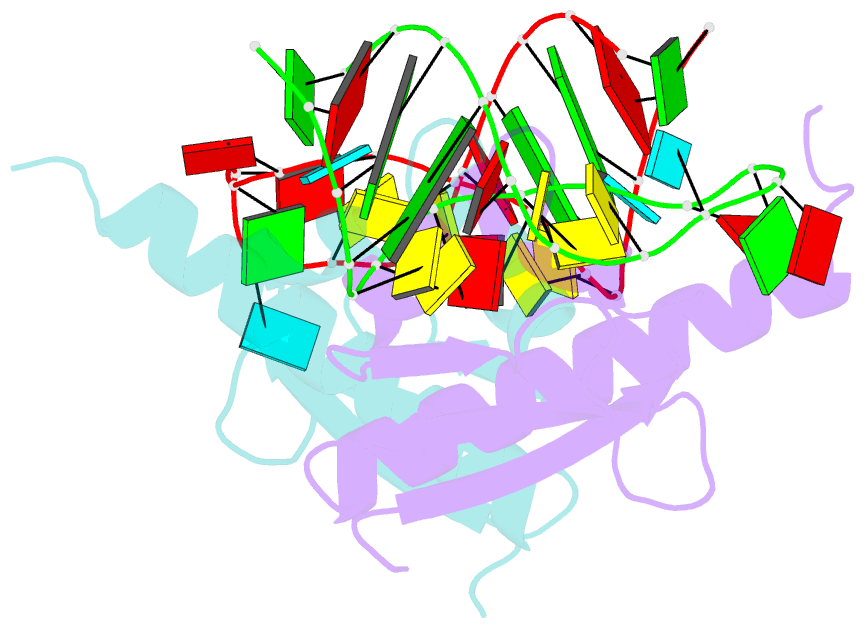
Summary information and primary citation
- PDB-id
-
1ec6;
DSSR-derived features in text and
JSON formats; DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.4 Å)
- Summary
- Crystal structure of nova-2 kh3 k-homology RNA-binding
domain bound to 20-mer RNA hairpin
- Reference
-
Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell
RB, Burley SK (2000): "Sequence-specific
RNA binding by a Nova KH domain: implications for
paraneoplastic disease and the fragile X syndrome."
Cell(Cambridge,Mass.), 100,
323-332. doi: 10.1016/S0092-8674(00)80668-6.
- Abstract
- The structure of a Nova protein K homology (KH) domain
recognizing single-stranded RNA has been determined at 2.4
A resolution. Mammalian Nova antigens (1 and 2) constitute
an important family of regulators of RNA metabolism in
neurons, first identified using sera from cancer patients
with the autoimmune disorder paraneoplastic
opsoclonus-myoclonus ataxia (POMA). The structure of the
third KH domain (KH3) of Nova-2 bound to a stem loop RNA
resembles a molecular vise, with 5'-Ura-Cyt-Ade-Cyt-3'
pinioned between an invariant Gly-X-X-Gly motif and the
variable loop. Tetranucleotide recognition is supported by
an aliphatic alpha helix/beta sheet RNA-binding platform,
which mimics 5'-Ura-Gua-3' by making Watson-Crick-like
hydrogen bonds with 5'-Cyt-Ade-3'. Sequence conservation
suggests that fragile X mental retardation results from
perturbation of RNA binding by the FMR1 protein.