Summary information and primary citation
- PDB-id
- 1efw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (3.0 Å)
- Summary
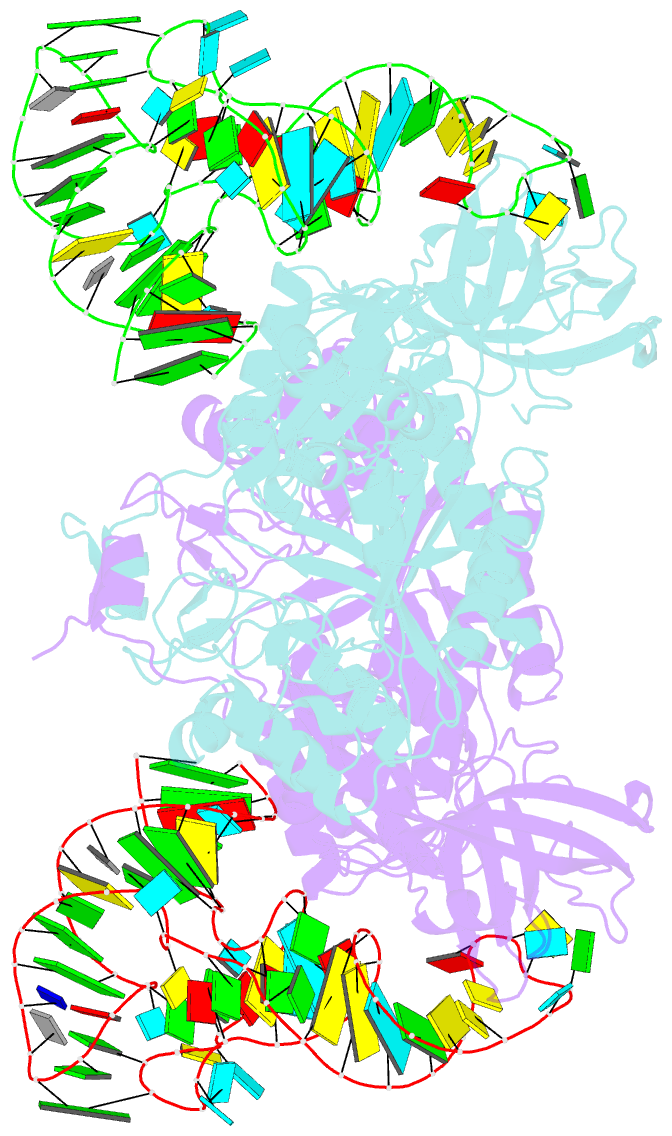
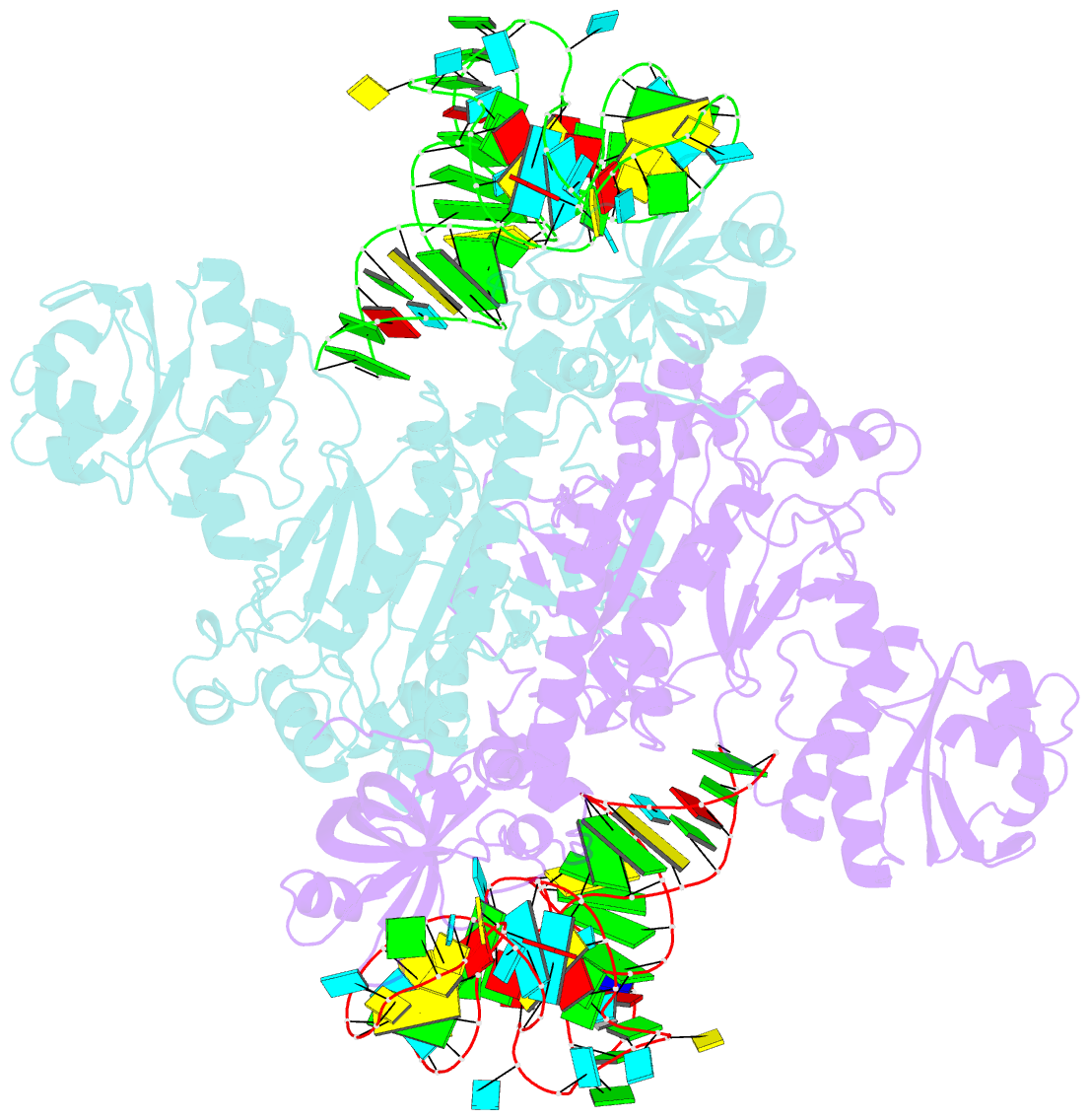
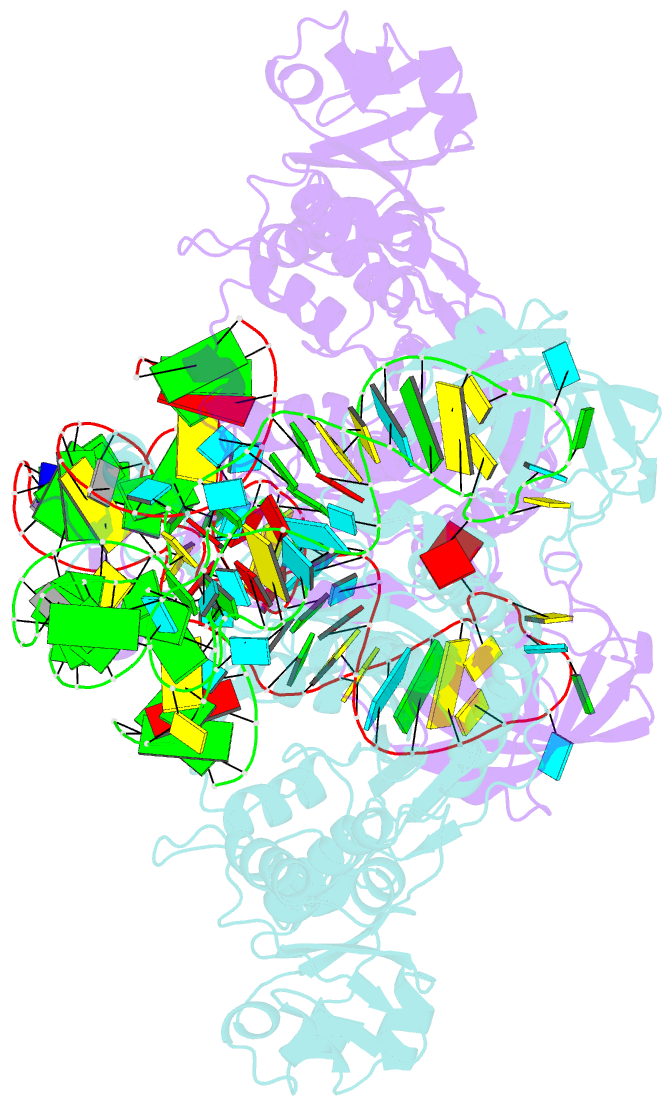
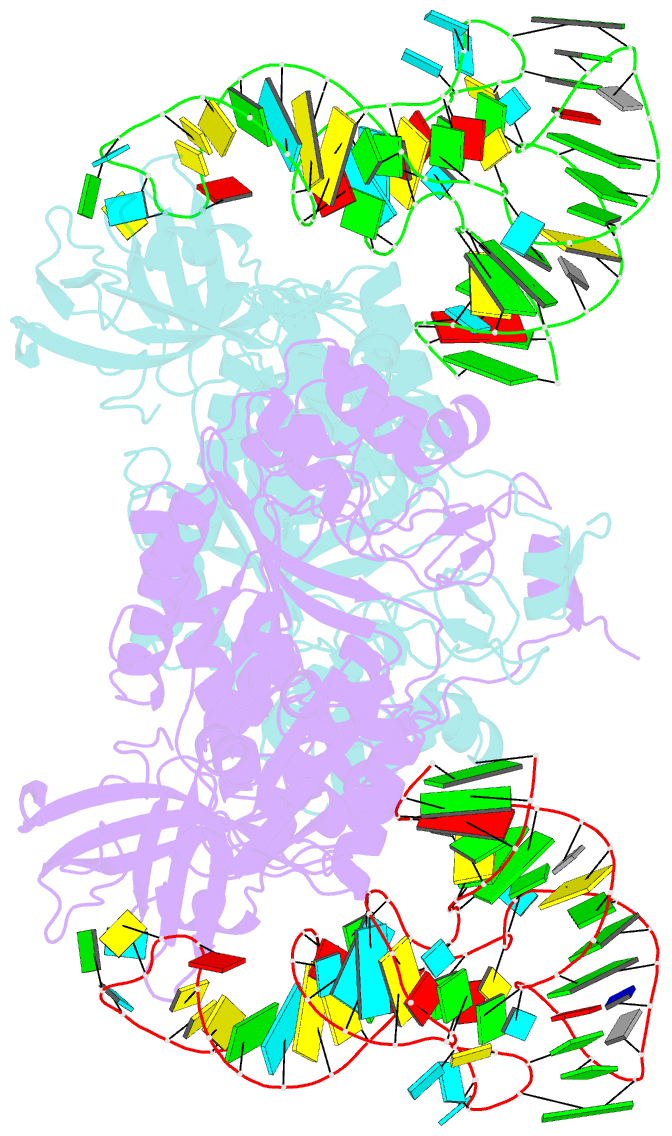
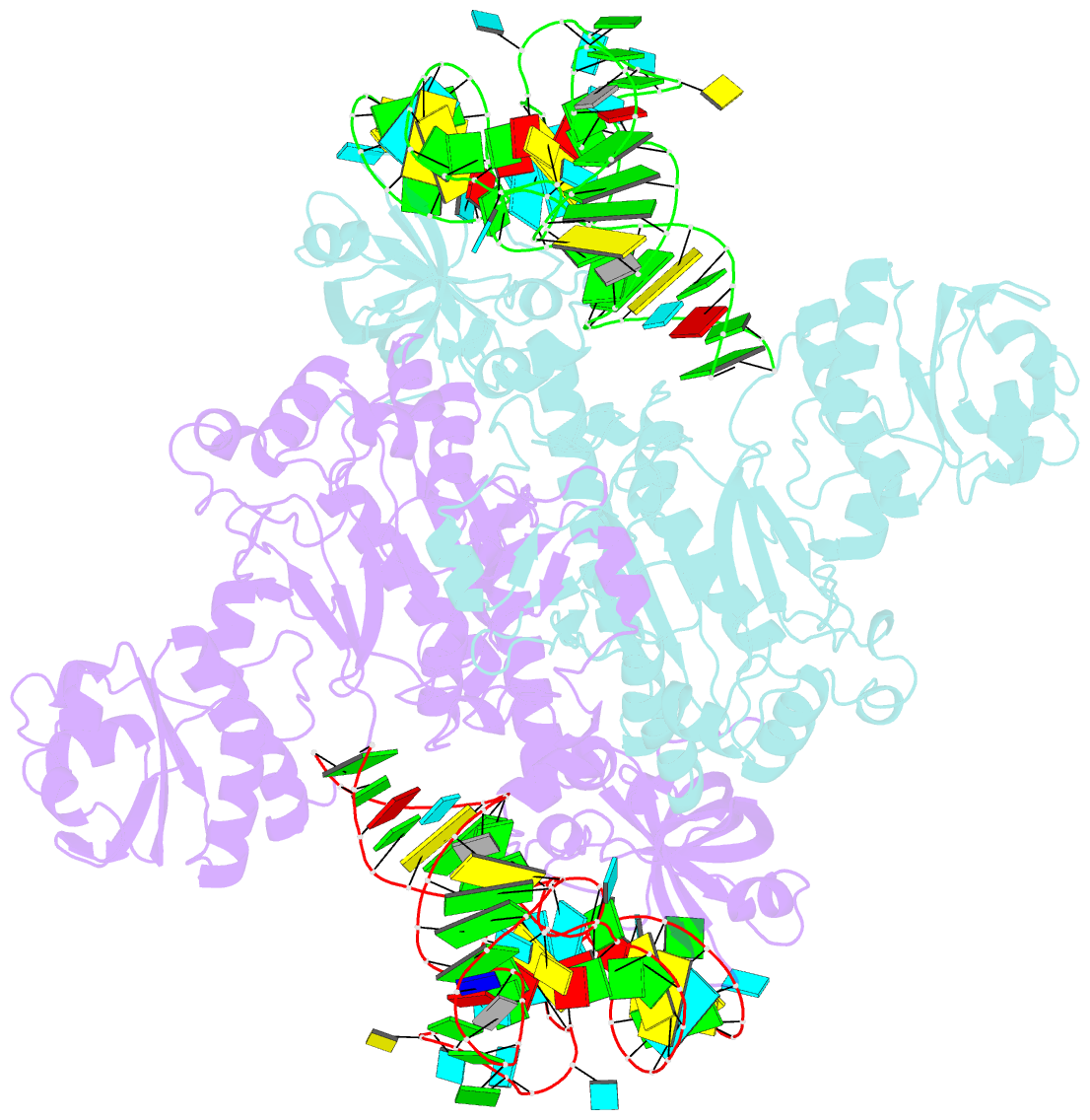
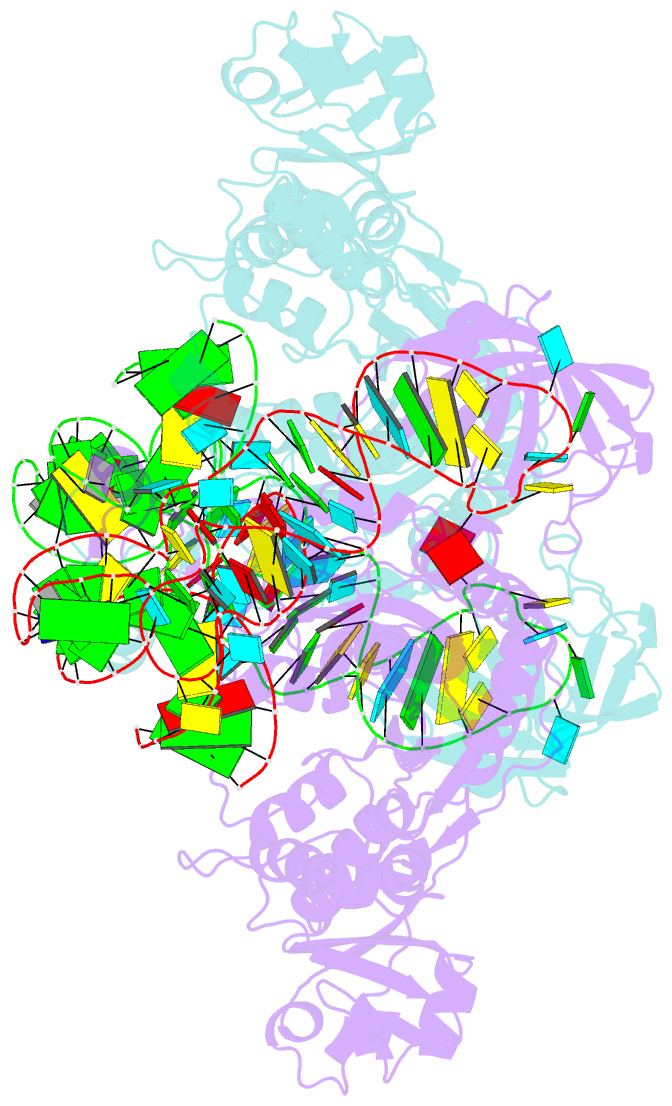
- Crystal structure of aspartyl-trna synthetase from thermus thermophilus complexed to trnaasp from escherichia coli
- Reference
- Briand C, Poterszman A, Eiler S, Webster G, Thierry J, Moras D (2000): "An intermediate step in the recognition of tRNA(Asp) by aspartyl-tRNA synthetase." J.Mol.Biol., 299, 1051-1060. doi: 10.1006/jmbi.2000.3819.
- Abstract
- The crystal structures of aspartyl-tRNA synthetase (AspRS) from Thermus thermophilus, a prokaryotic class IIb enzyme, complexed with tRNA(Asp) from either T. thermophilus or Escherichia coli reveal a potential intermediate of the recognition process. The tRNA is positioned on the enzyme such that it cannot be aminoacylated but adopts an overall conformation similar to that observed in active complexes. While the anticodon loop binds to the N-terminal domain of the enzyme in a manner similar to that of the related active complexes, its aminoacyl acceptor arm remains at the entrance of the active site, stabilized in its intermediate conformational state by non-specific interactions with the insertion and catalytic domains. The thermophilic nature of the enzyme, which manifests itself in a very low kinetic efficiency at 17 degrees C, the temperature at which the crystals were grown, is in agreement with the relative stability of this non-productive conformational state. Based on these data, a pathway for tRNA binding and recognition is proposed.