Summary information and primary citation
- PDB-id
- 1ekz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- cell cycle-RNA
- Method
- NMR
- Summary
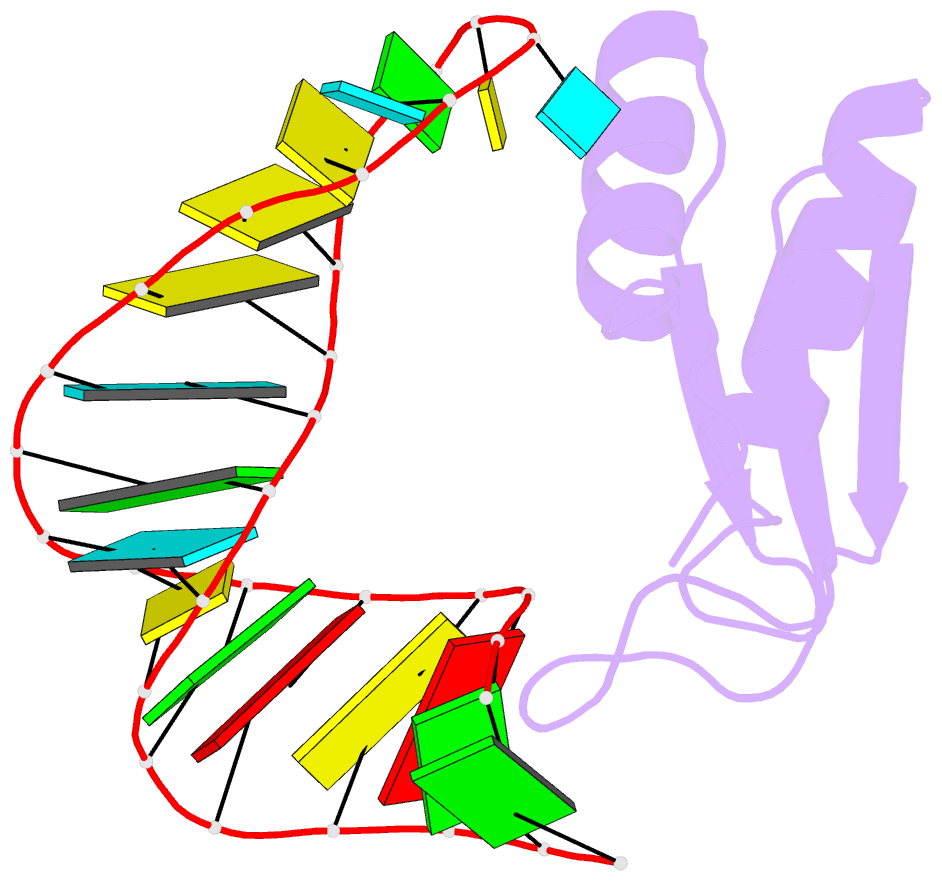
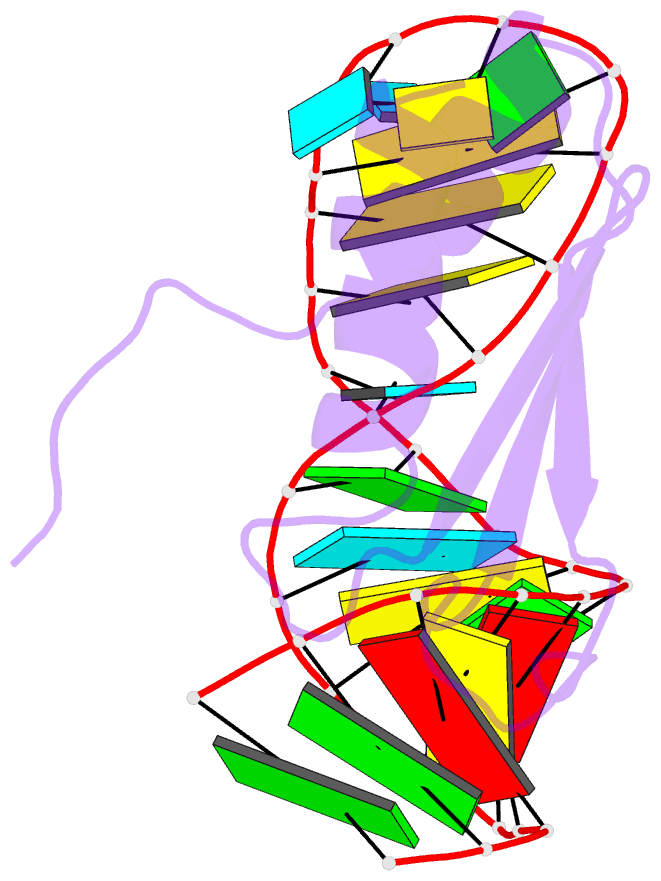
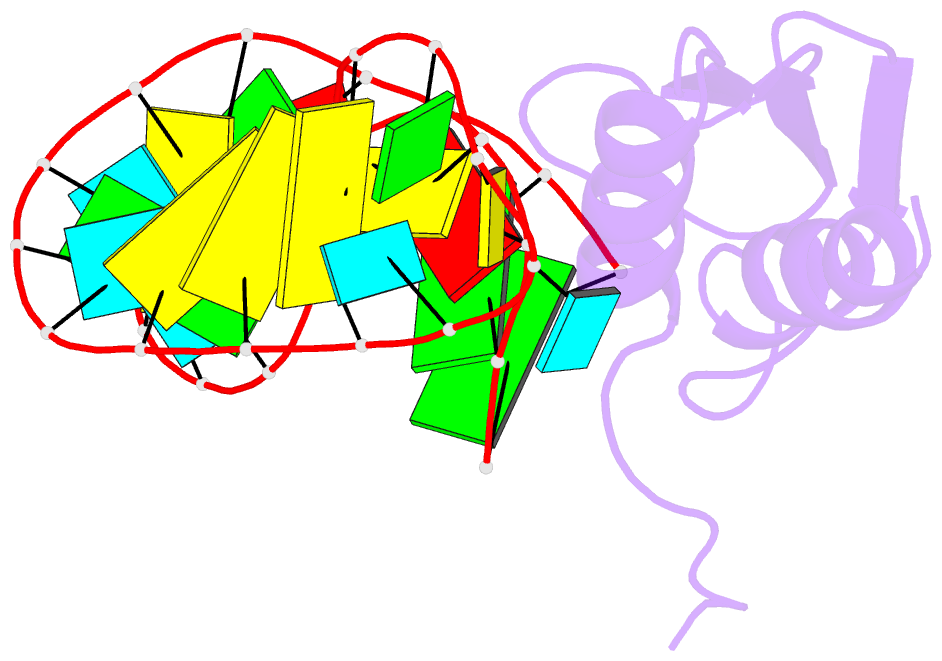
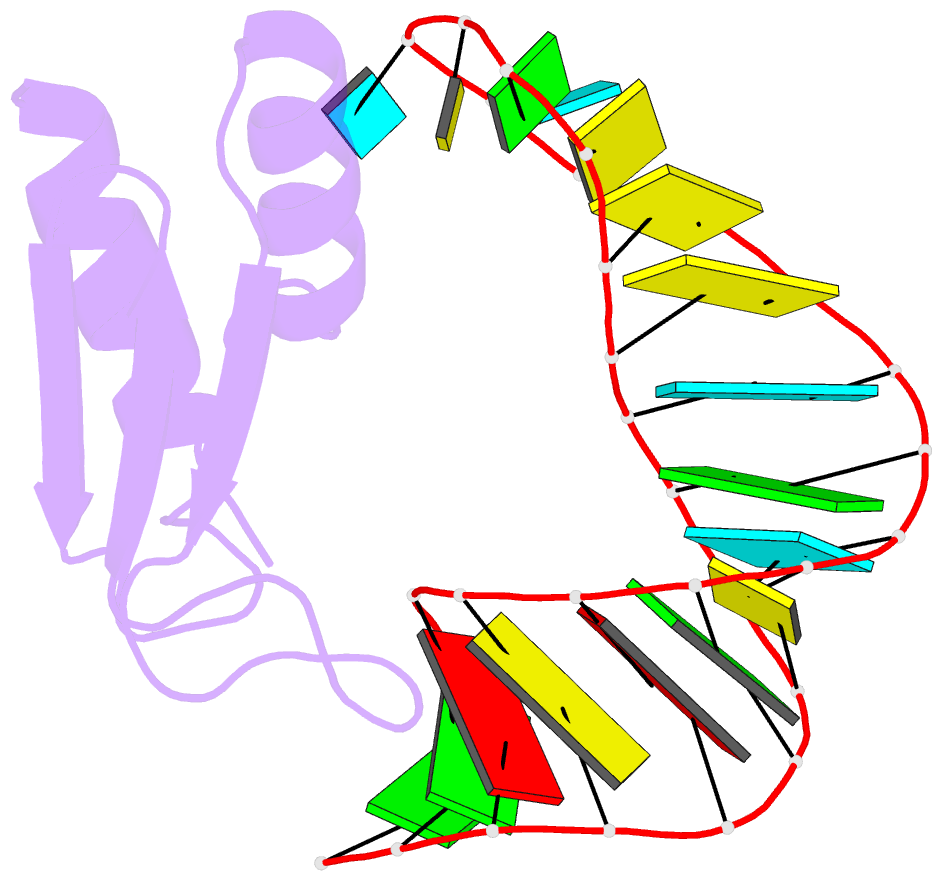
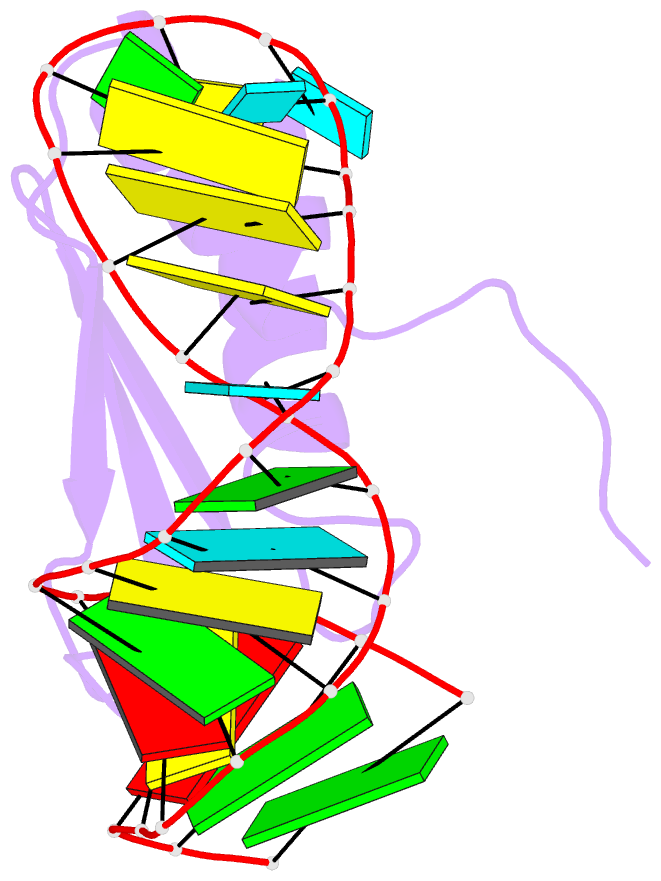
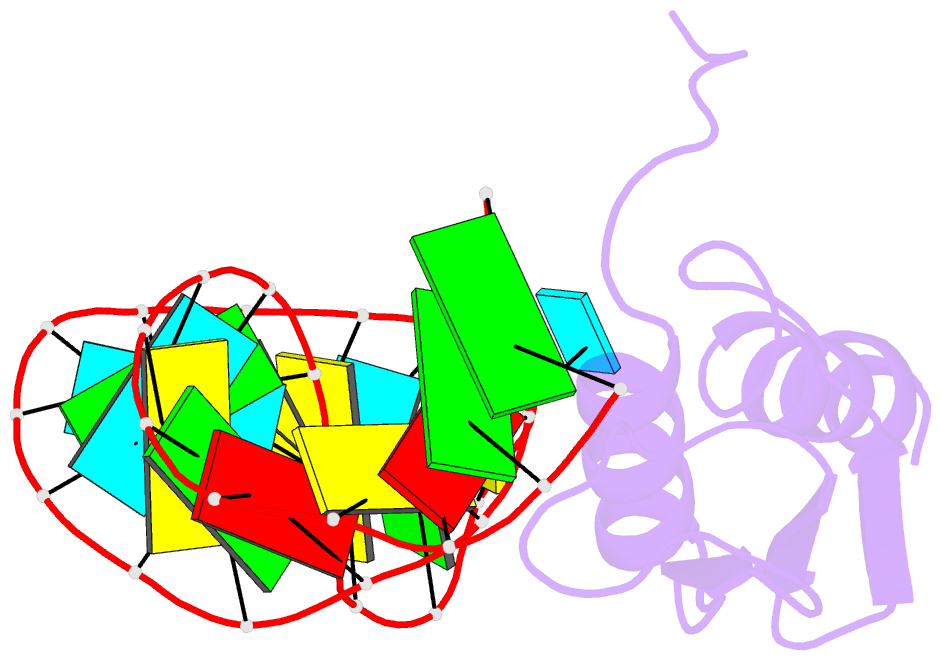
- NMR structure of the complex between the third dsrbd from drosophila staufen and a RNA hairpin
- Reference
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G (2000): "RNA recognition by a Staufen double-stranded RNA-binding domain." EMBO J., 19, 997-1009. doi: 10.1093/emboj/19.5.997.
- Abstract
- The double-stranded RNA-binding domain (dsRBD) is a common RNA-binding motif found in many proteins involved in RNA maturation and localization. To determine how this domain recognizes RNA, we have studied the third dsRBD from Drosophila Staufen. The domain binds optimally to RNA stem-loops containing 12 uninterrupted base pairs, and we have identified the amino acids required for this interaction. By mutating these residues in a staufen transgene, we show that the RNA-binding activity of dsRBD3 is required in vivo for Staufen-dependent localization of bicoid and oskar mRNAs. Using high-resolution NMR, we have determined the structure of the complex between dsRBD3 and an RNA stem-loop. The dsRBD recognizes the shape of A-form dsRNA through interactions between conserved residues within loop 2 and the minor groove, and between loop 4 and the phosphodiester backbone across the adjacent major groove. In addition, helix alpha1 interacts with the single-stranded loop that caps the RNA helix. Interactions between helix alpha1 and single-stranded RNA may be important determinants of the specificity of dsRBD proteins.