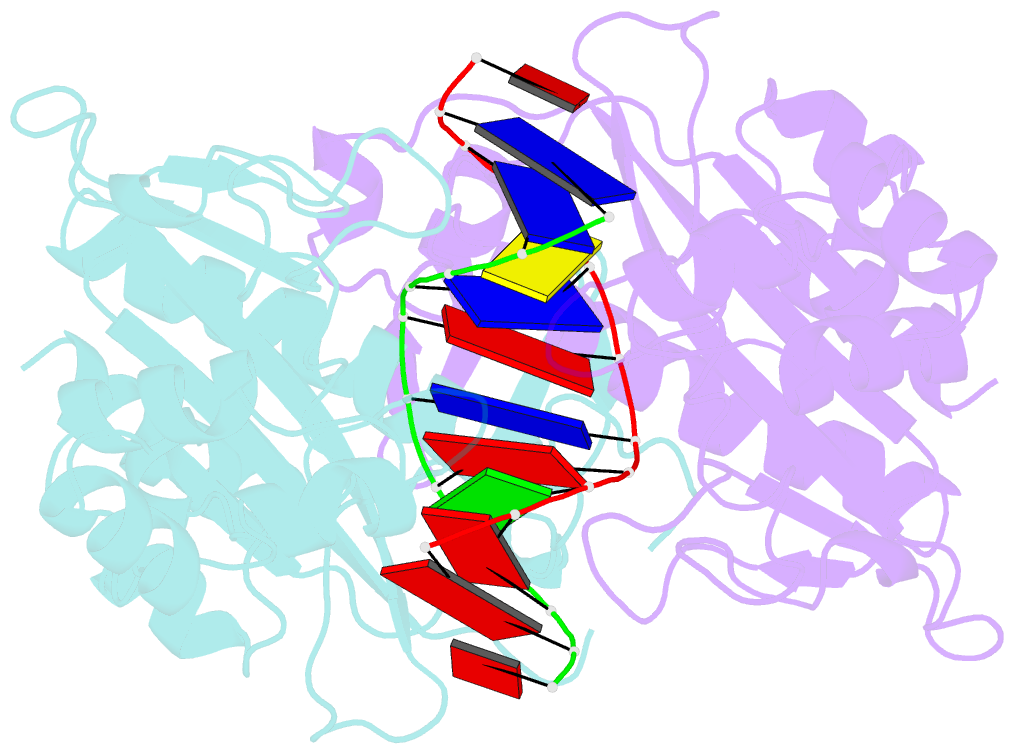
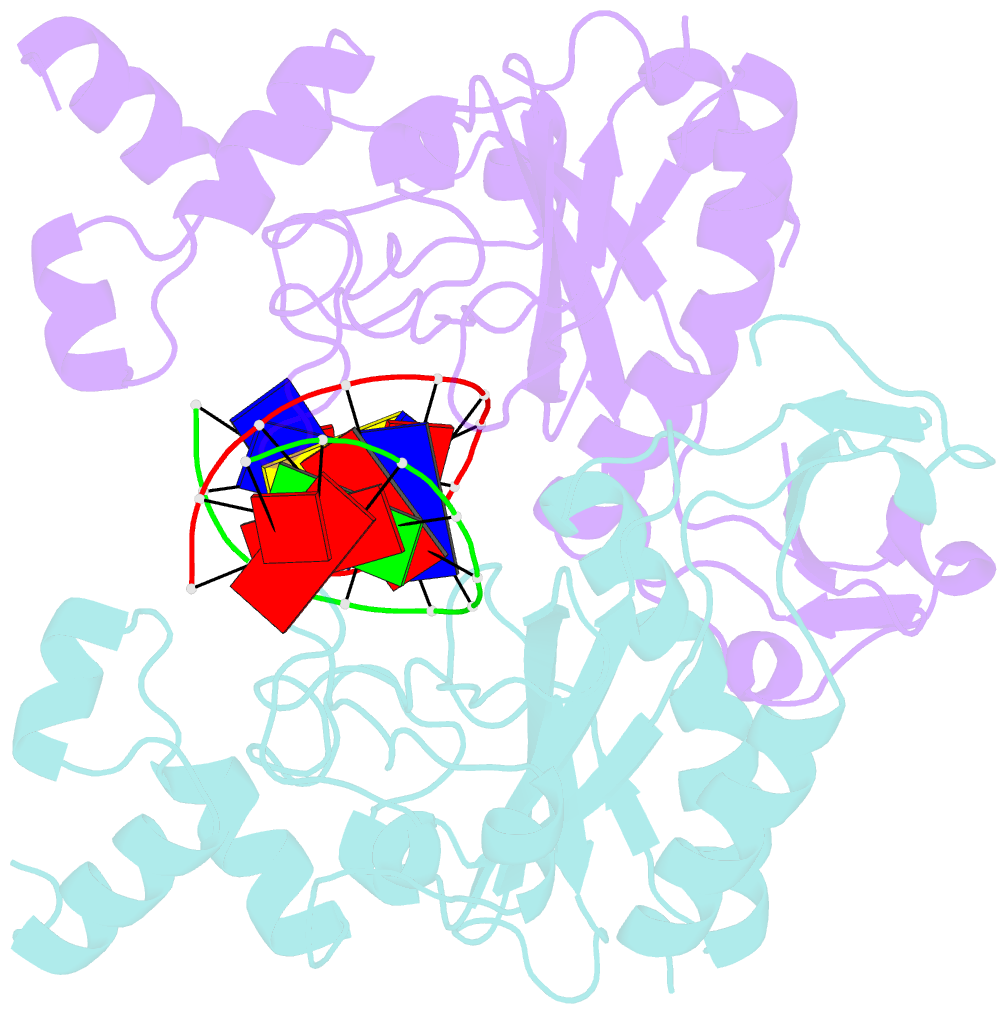
Summary information and primary citation
- PDB-id
- 1eop; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.6 Å)
- Summary
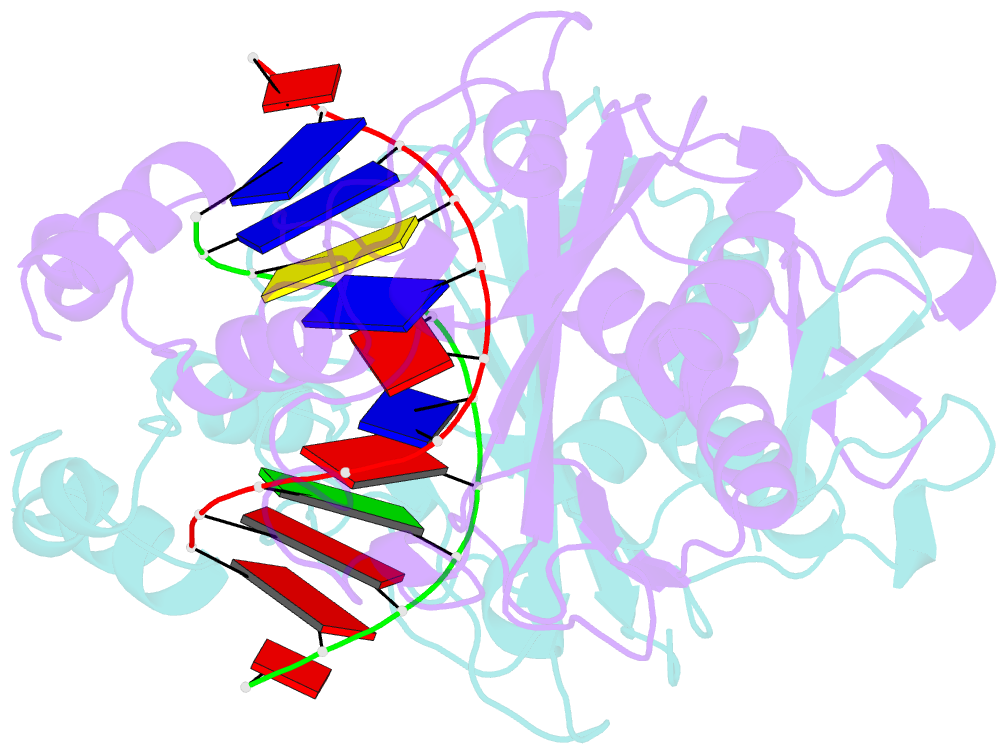
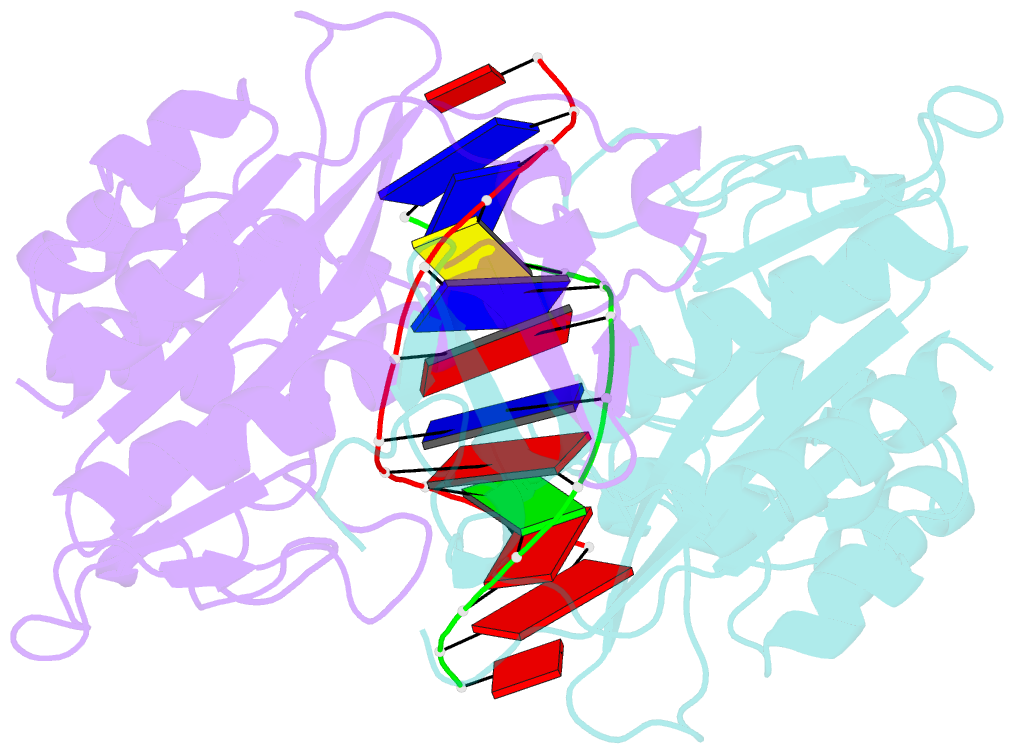
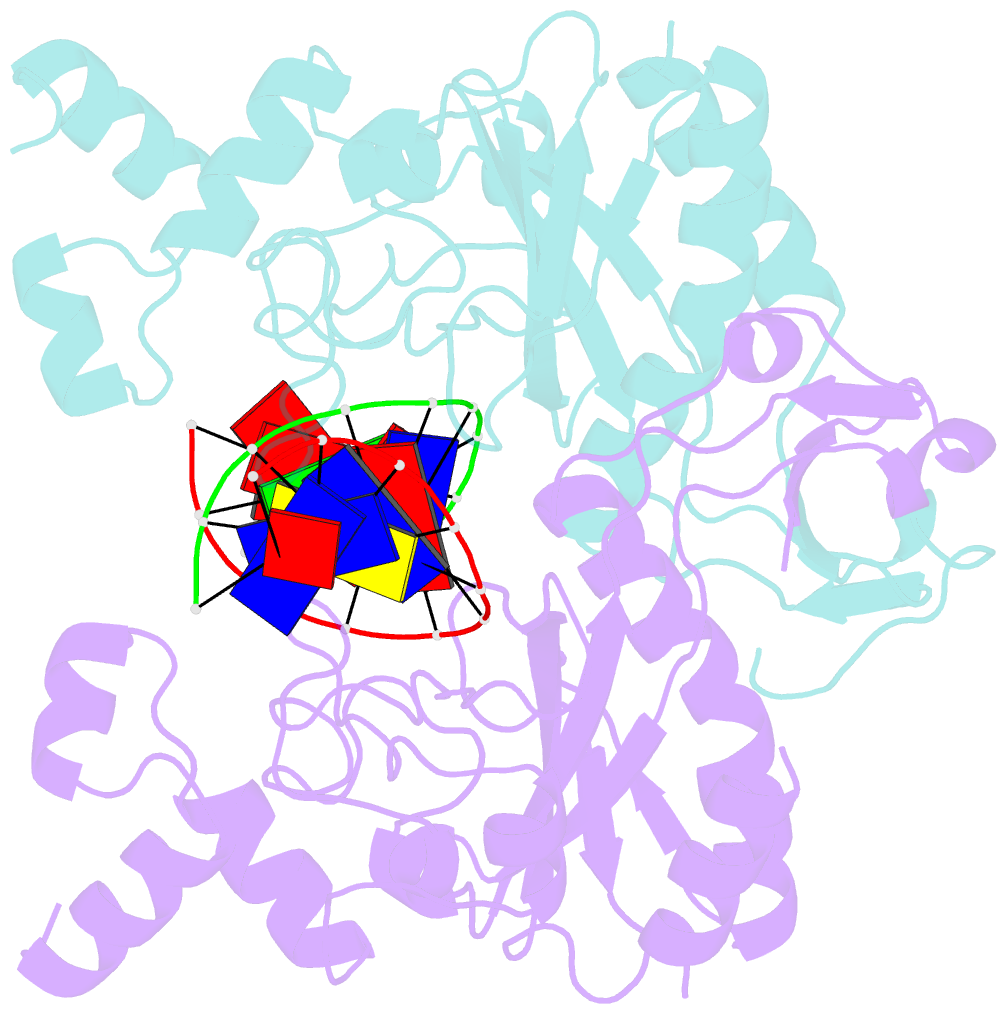
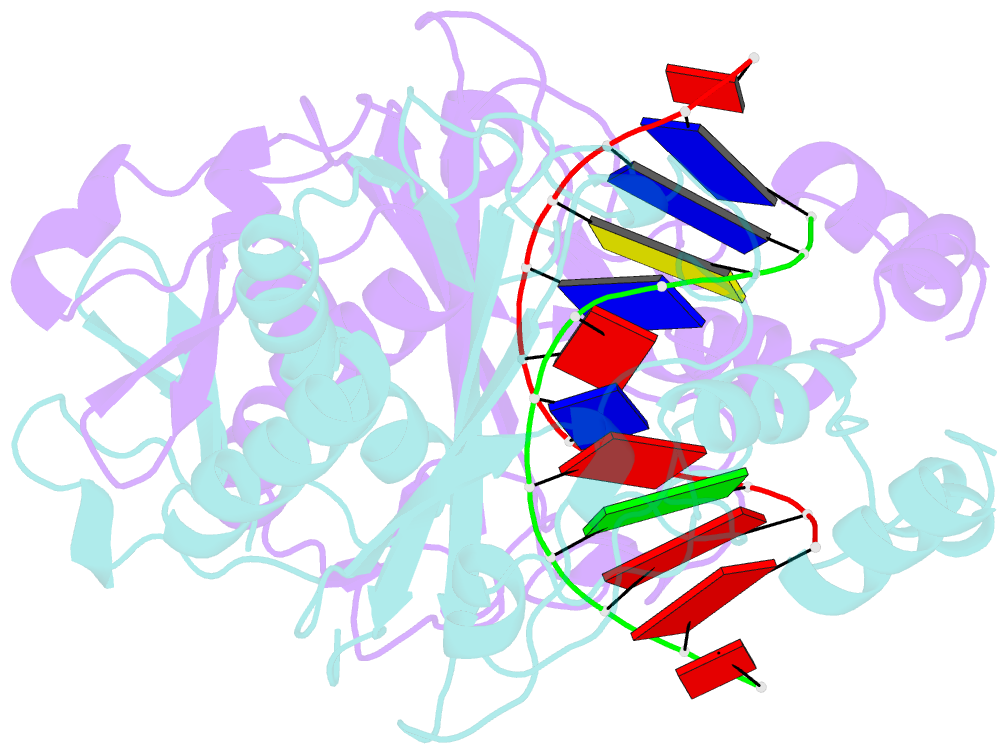
- Ecorv bound to cognate DNA
- Reference
- Horton NC, Perona JJ (2000): "Crystallographic snapshots along a protein-induced DNA-bending pathway." Proc.Natl.Acad.Sci.USA, 97, 5729-5734. doi: 10.1073/pnas.090370797.
- Abstract
- Two new high-resolution cocrystal structures of EcoRV endonuclease bound to DNA show that a large variation in DNA-bending angles is sampled in the ground state binary complex. Together with previous structures, these data reveal a contiguous series of protein conformational states delineating a specific trajectory for the induced-fit pathway. Rotation of the DNA-binding domains, together with movements of two symmetry-related helices binding in the minor groove, causes base unstacking at a key base-pair step and propagates structural changes that assemble the active sites. These structures suggest a complex mechanism for DNA bending that depends on forces generated by interacting protein segments, and on selective neutralization of phosphate charges along the inner face of the bent double helix.