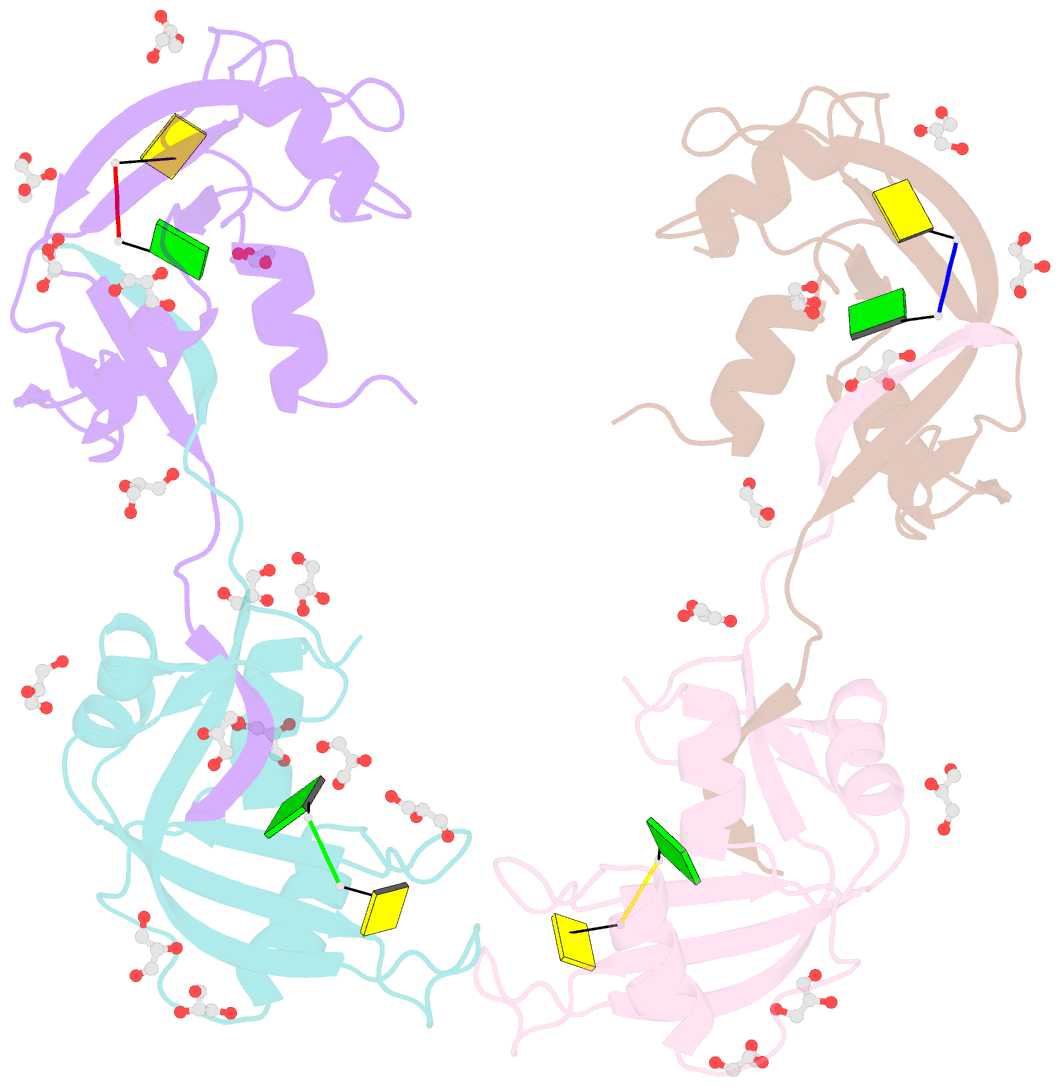
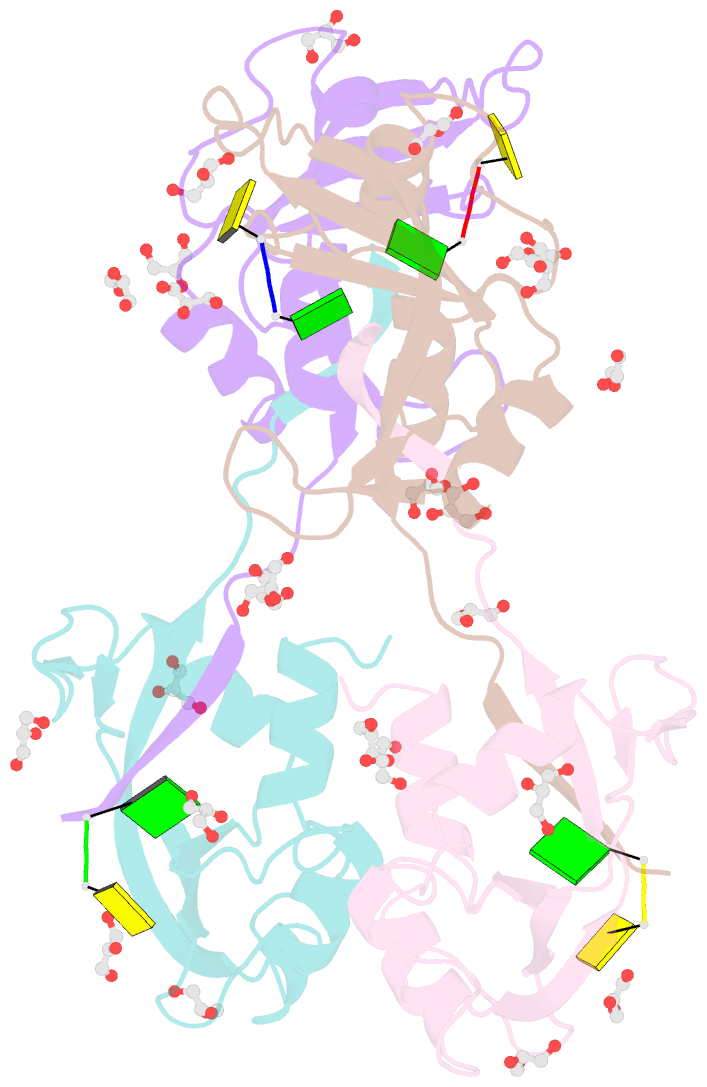
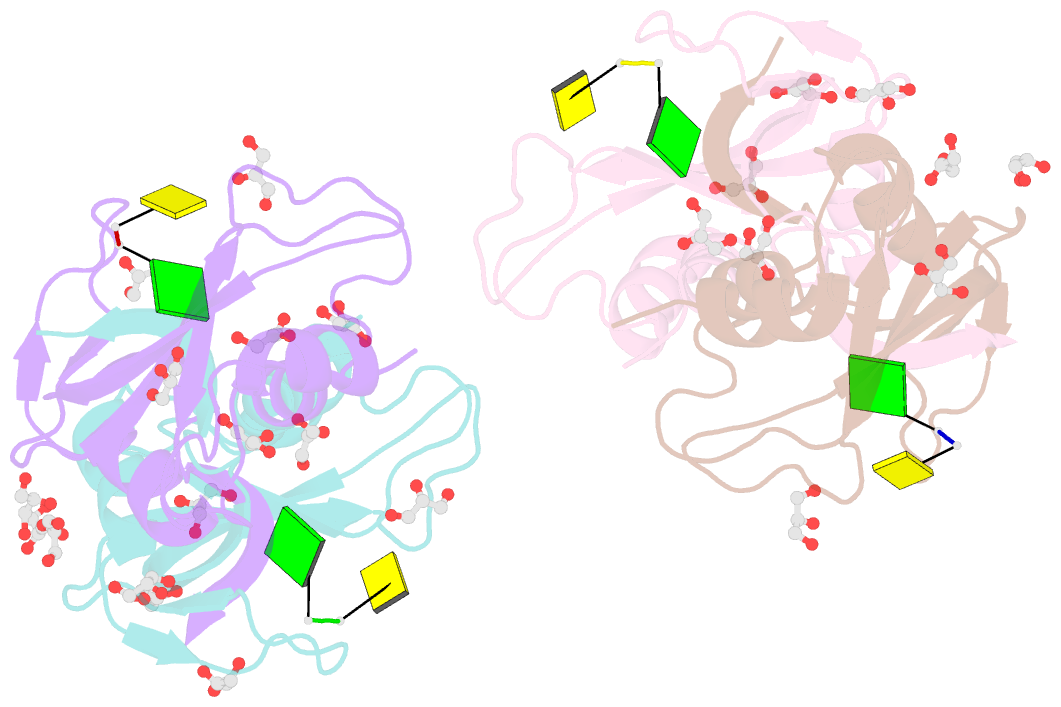
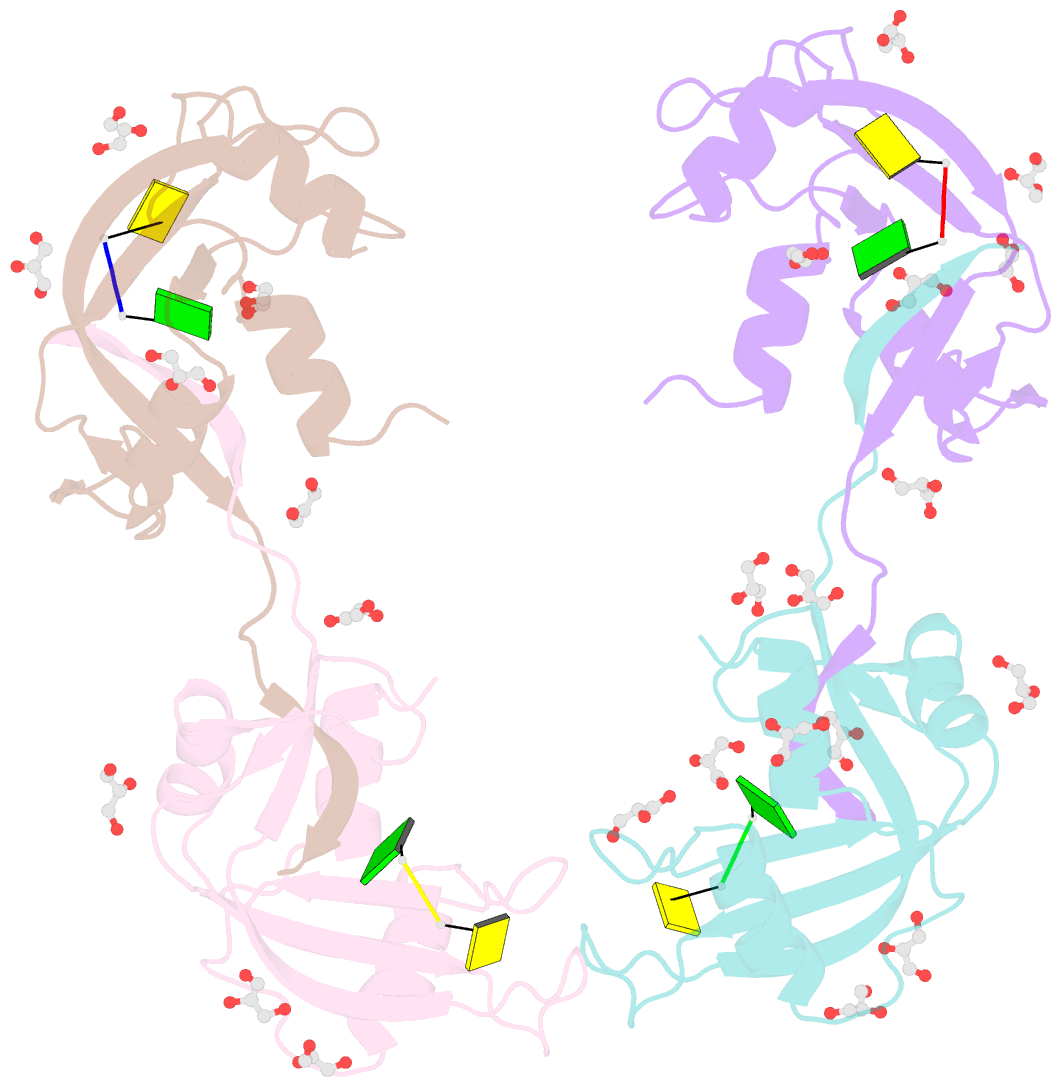
Summary information and primary citation
- PDB-id
- 1f0v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.7 Å)
- Summary
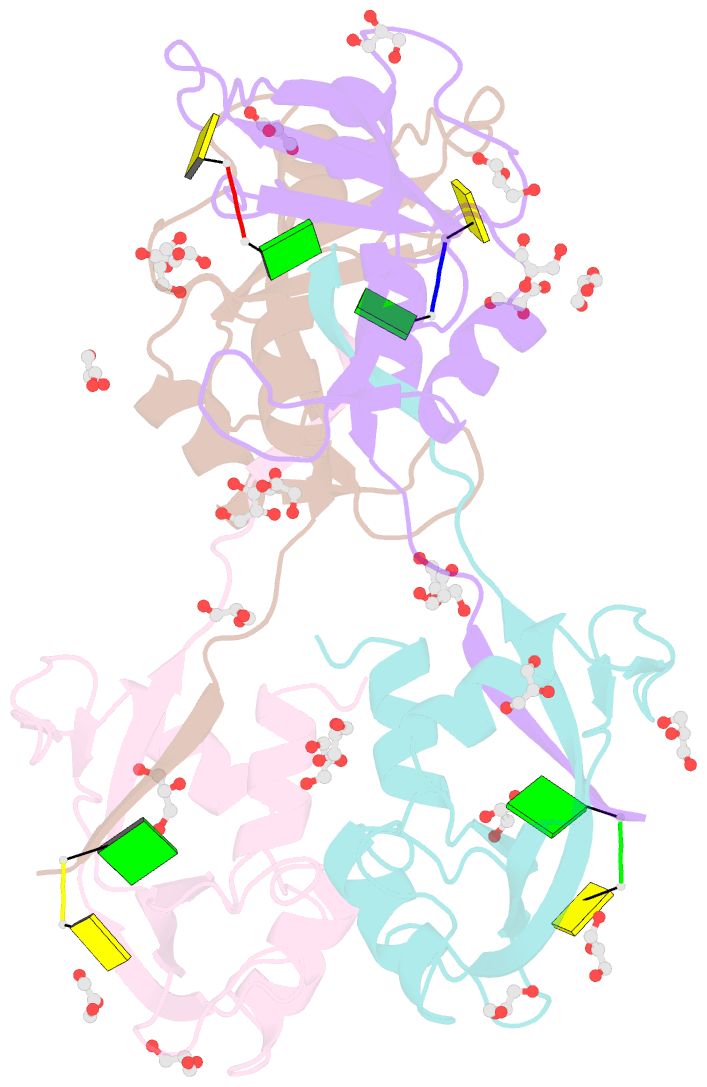
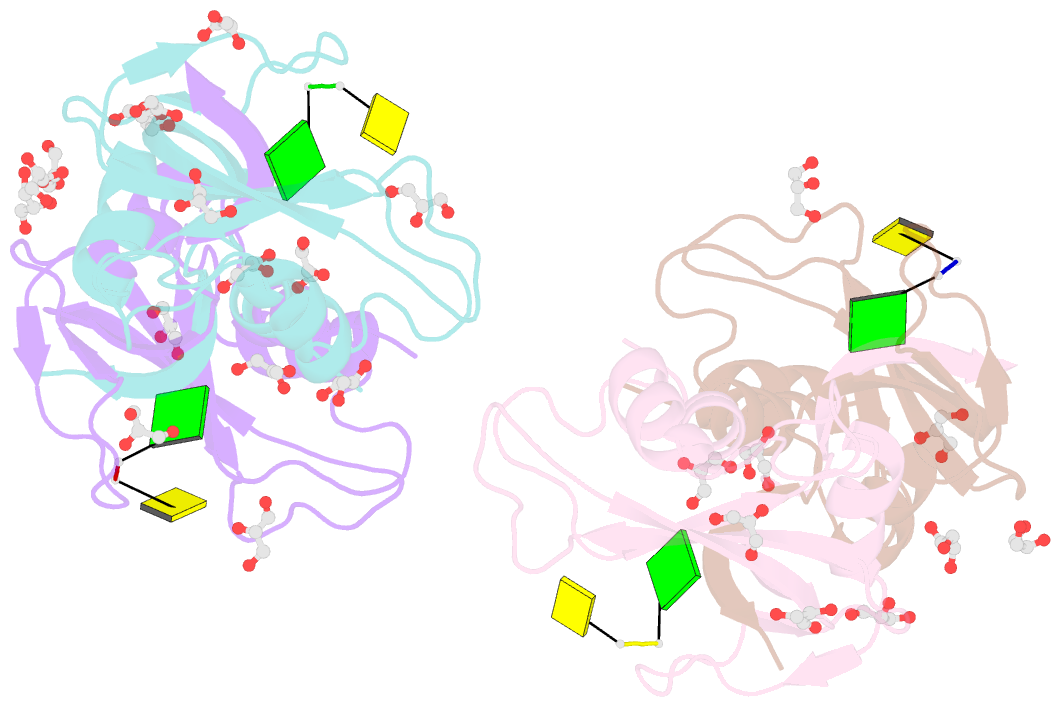
- Crystal structure of an rnase a dimer displaying a new type of 3d domain swapping
- Reference
- Liu YS, Gotte G, Libonati M, Eisenberg DS (2001): "A domain-swapped RNase A dimer with implications for amyloid formation." Nat.Struct.Biol., 8, 211-214. doi: 10.1038/84941.
- Abstract
- Bovine pancreatic ribonuclease (RNase A) forms two types of dimers (a major and a minor component) upon concentration in mild acid. These two dimers exhibit different biophysical and biochemical properties. Earlier we reported that the minor dimer forms by swapping its N-terminal alpha-helix with that of an identical molecule. Here we find that the major dimer forms by swapping its C-terminal beta-strand, thus revealing the first example of three-dimensional (3D) domain swapping taking place in different parts of the same protein. This feature permits RNase A to form tightly bonded higher oligomers. The hinge loop of the major dimer, connecting the swapped beta-strand to the protein core, resembles a short segment of the polar zipper proposed by Perutz and suggests a model for aggregate formation by 3D domain swapping with a polar zipper.