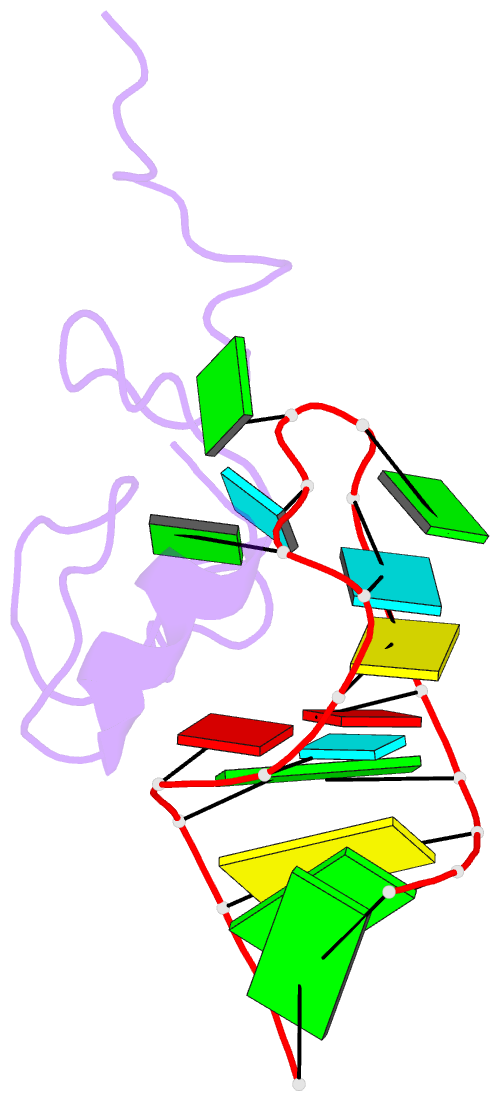
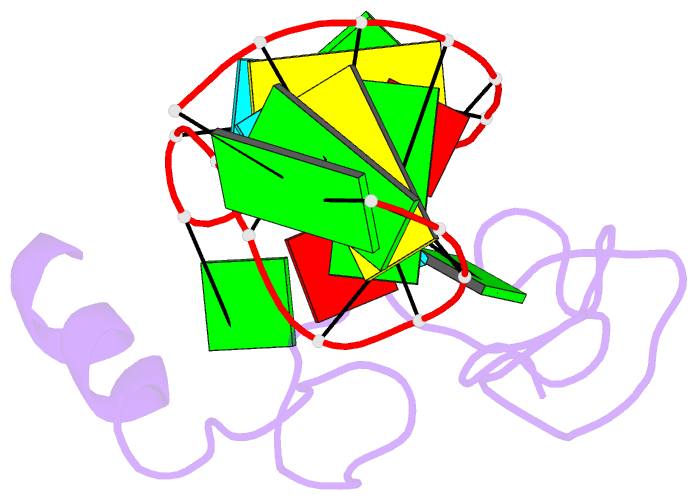
Summary information and primary citation
- PDB-id
- 1f6u; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-RNA
- Method
- NMR
- Summary
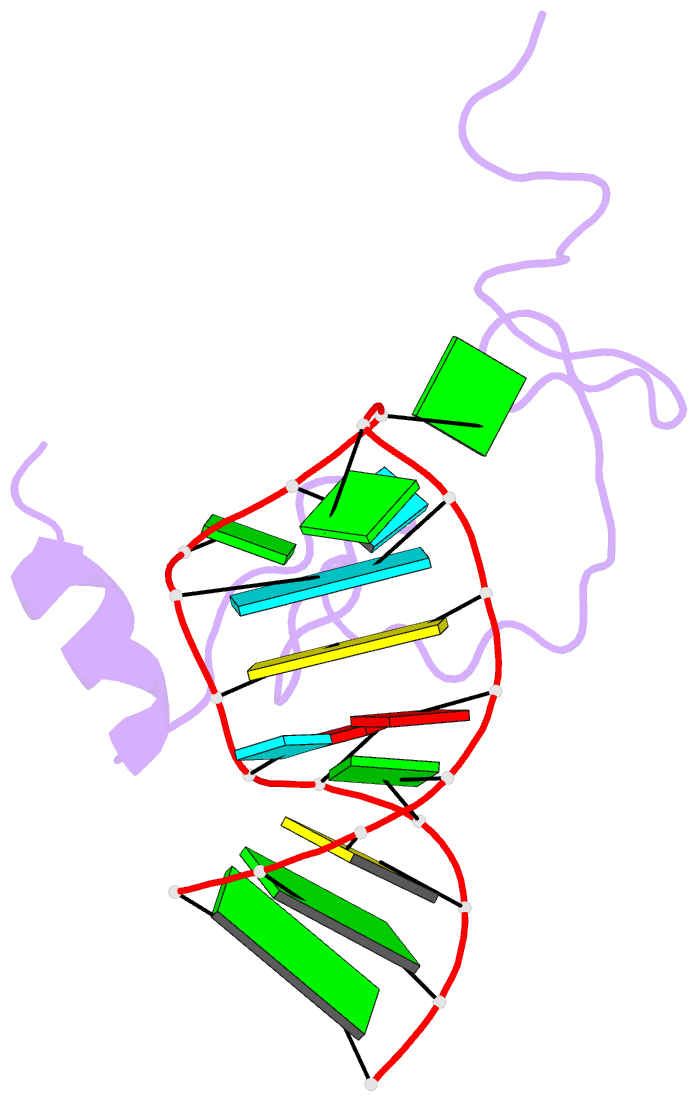
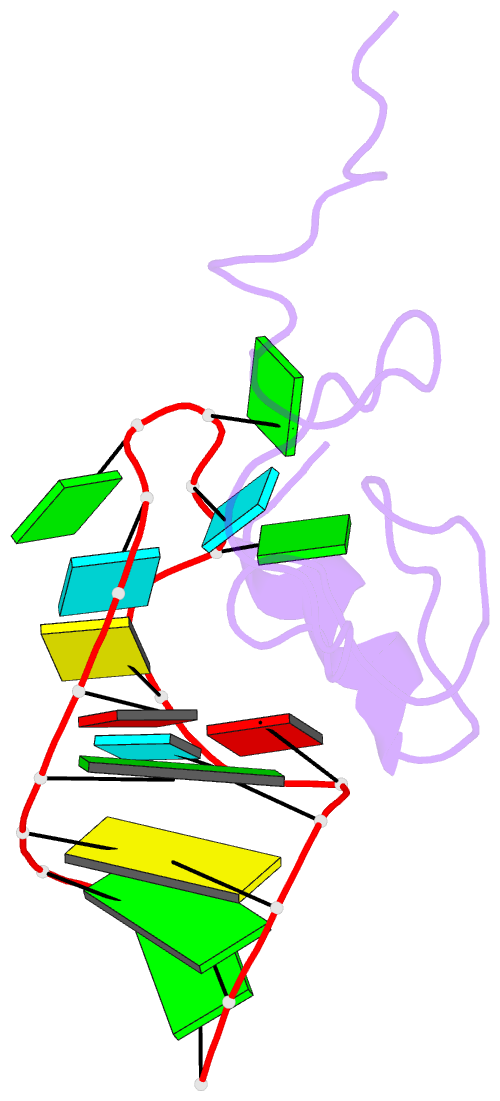
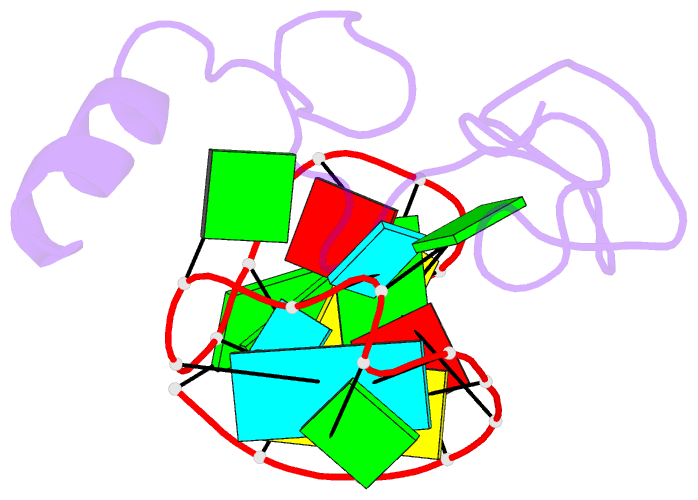
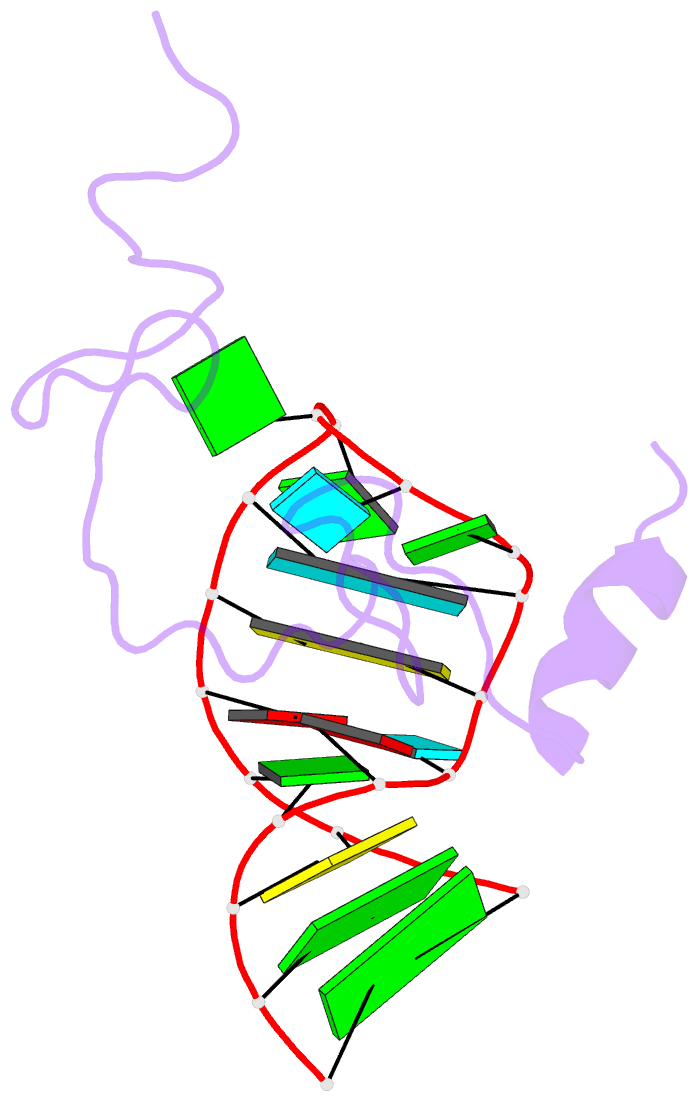
- NMR structure of the hiv-1 nucleocapsid protein bound to stem-loop sl2 of the psi-RNA packaging signal. implications for genome recognition
- Reference
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF (2000): "NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition." J.Mol.Biol., 301, 491-511. doi: 10.1006/jmbi.2000.3979.
- Abstract
- The RNA genome of the human immunodeficiency virus type-1 (HIV-1) contains a approximately 120 nucleotide Psi-packaging signal that is recognized by the nucleocapsid (NC) domain of the Gag polyprotein during virus assembly. The Psi-site contains four stem-loops (SL1-SL4) that possess overlapping and possibly redundant functions. The present studies demonstrate that the 19 residue SL2 stem-loop binds NC with affinity (K(d)=110(+/-50) nM) similar to that observed for NC binding to SL3 (K(d)=170(+/-65) nM) and tighter than expected on the basis of earlier work, suggesting that NC-SL2 interactions probably play a direct role in the specific recognition and packaging of the full-length, unspliced genome. The structure of the NC-SL2 complex was determined by heteronuclear NMR methods using (15)N,(13)C-isotopically labeled NC protein and SL2 RNA. The N and C-terminal "zinc knuckles" (Cys-X(2)-Cys-X(4)-His-X(4)-Cys; X=variable amino acid) of HIV-1 NC bind to exposed guanosine bases G9 and G11, respectively, of the G8-G9-U10-G11 tetraloop, and residues Lys3-Lys11 of the N-terminal tail forms a 3(10) helix that packs against the proximal zinc knuckle and interacts with the RNA stem. These structural features are similar to those observed previously in the NMR structure of NC bound to SL3. Other features of the complex are substantially different. In particular, the N-terminal zinc knuckle interacts with an A-U-A base triple platform in the minor groove of the SL2 RNA stem, but binds to the major groove of SL3. In addition, the relative orientations of the N and C-terminal zinc knuckles differ in the NC-SL2 and NC-SL3 complexes, and the side-chain of Phe6 makes minor groove hydrophobic contacts with G11 in the NC-SL2 complex but does not interact with RNA in the NC-SL3 complex. Finally, the N-terminal helix of NC interacts with the phosphodiester backbone of the SL2 RNA stem mainly via electrostatic interactions, but does not bind in the major groove or make specific H-bonding contacts as observed in the NC-SL3 structure. These findings demonstrate that NC binds in an adaptive manner to SL2 and SL3 via different subsets of inter and intra-molecular interactions, and support a genome recognition/packaging mechanism that involves interactions of two or more NC domains of assembling HIV-1 Gag molecules with multiple Psi-site stem-loop packaging elements during the early stages of retrovirus assembly.