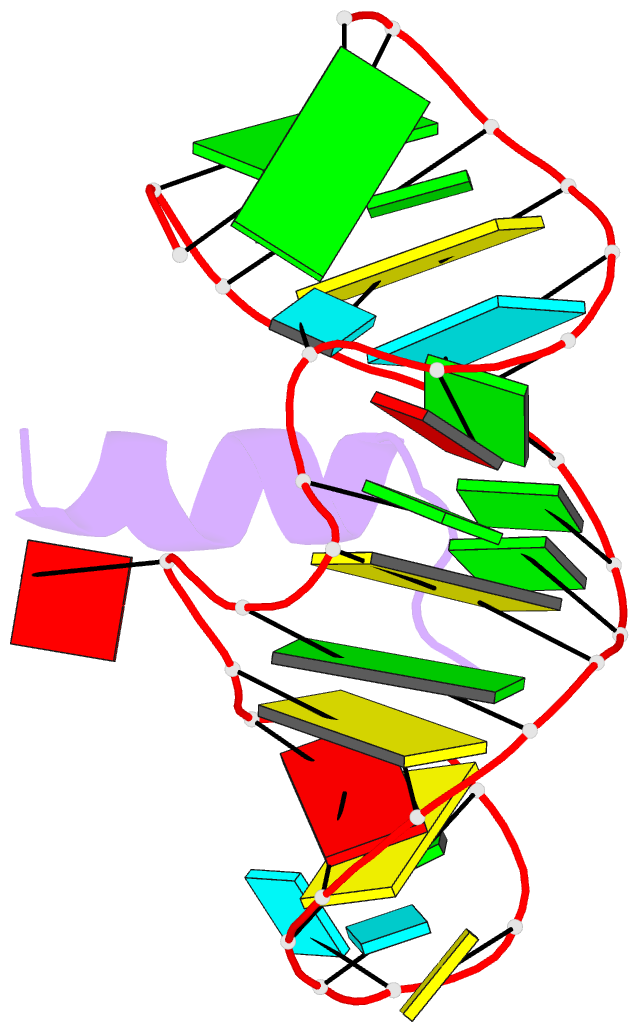
Summary information and primary citation
- PDB-id
- 1g70; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- NMR
- Summary
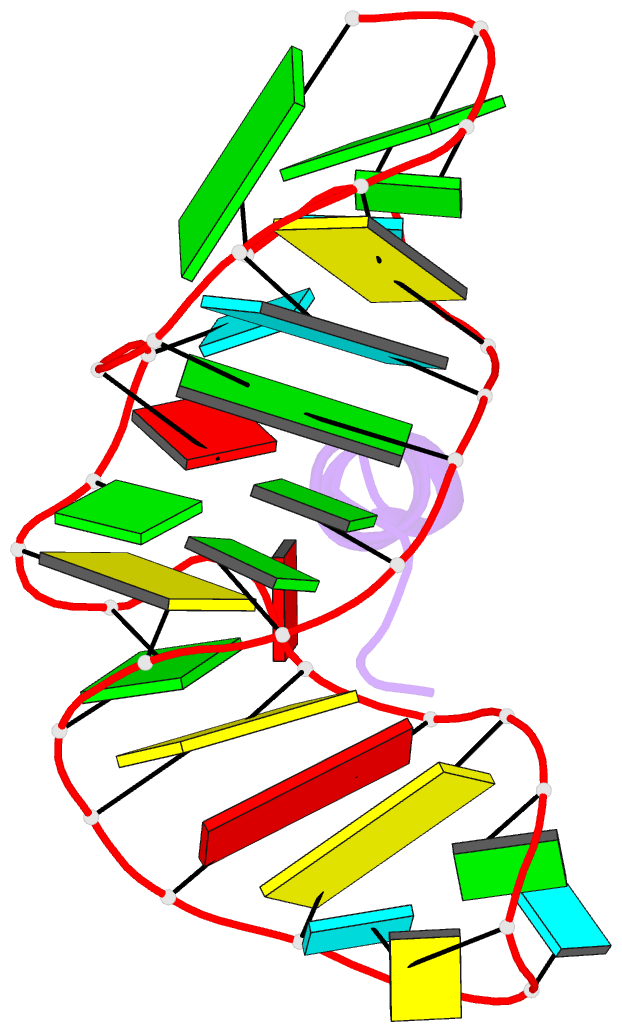
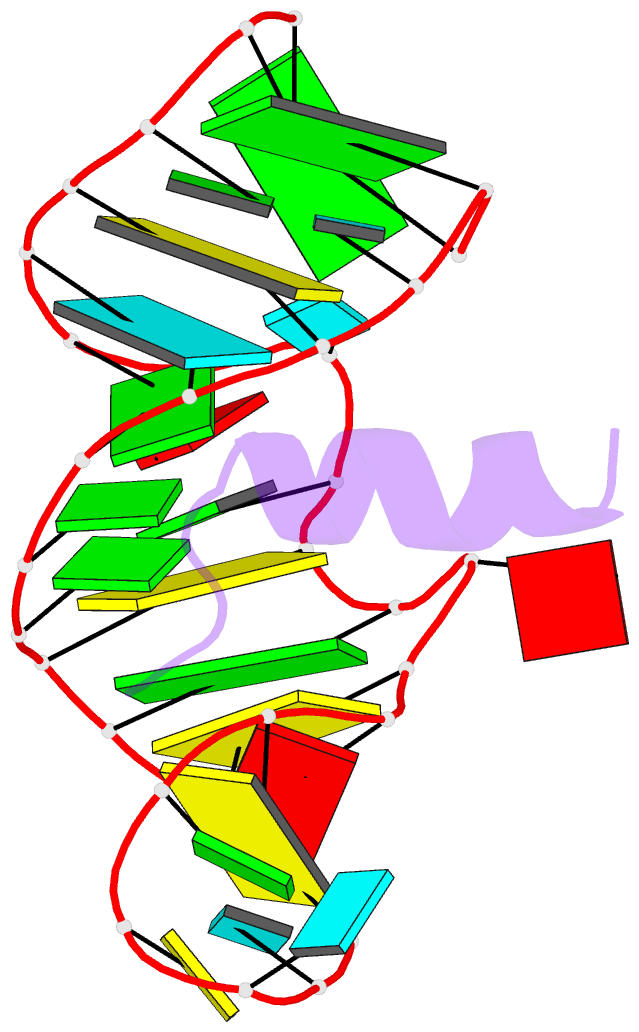
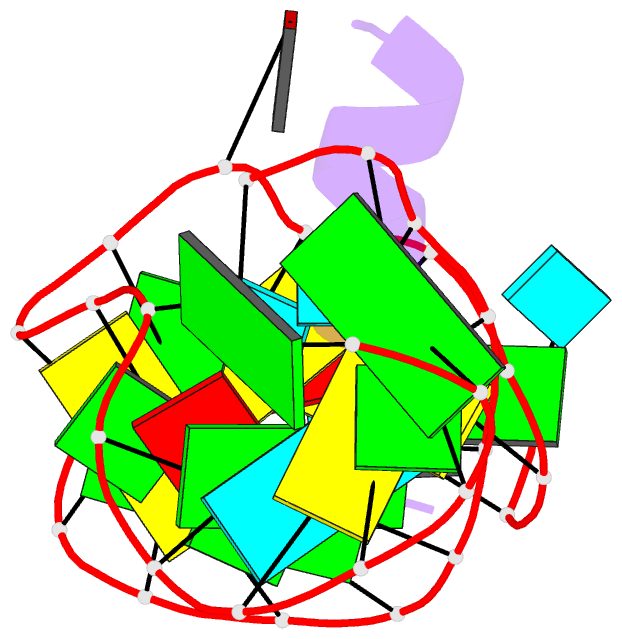
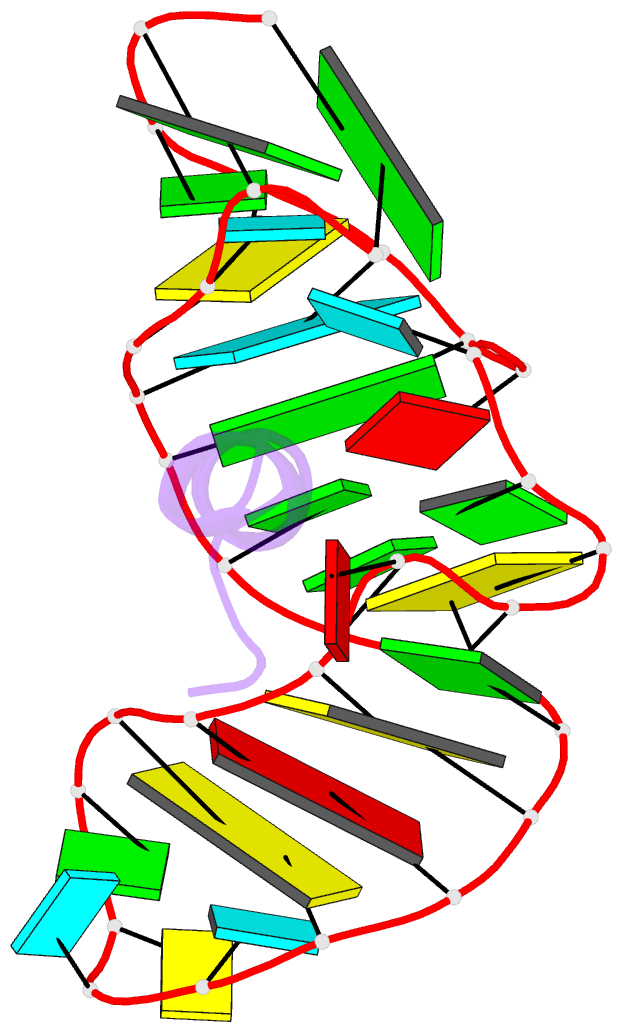
- Complex of hiv-1 rre-iib RNA with rsg-1.2 peptide
- Reference
- Gosser Y, Hermann T, Majumdar A, Hu W, Frederick R, Jiang F, Xu W, Patel DJ (2001): "Peptide-triggered conformational switch in HIV-1 RRE RNA complexes." Nat.Struct.Biol., 8, 146-150. doi: 10.1038/84138.
- Abstract
- We have used NMR spectroscopy to determine the solution structure of a complex between an oligonucleotide derived from stem IIB of the Rev responsive element (RRE-IIB) of HIV-1 mRNA and an in vivo selected, high affinity binding Arg-rich peptide. The peptide binds in a partially alpha-helical conformation into a pocket within the RNA deep groove. Comparison with the structure of a complex between an alpha-helical Rev peptide and RRE-IIB reveals that the sequence of the bound peptide determines the local conformation of the RRE peptide binding site. A conformational switch of an unpaired uridine base was revealed; this points out into the solvent in the Rev peptide complex, but it is stabilized inside the RNA deep groove by stacking with an Arg side chain in the selected peptide complex. The conformational switch has been visualized by NMR chemical shift mapping of the uridine H5/H6 atoms during a competition experiment in which Rev peptide was displaced from RRE-IIB by the higher affinity binding selected peptide.