Summary information and primary citation
- PDB-id
- 1gd2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.0 Å)
- Summary
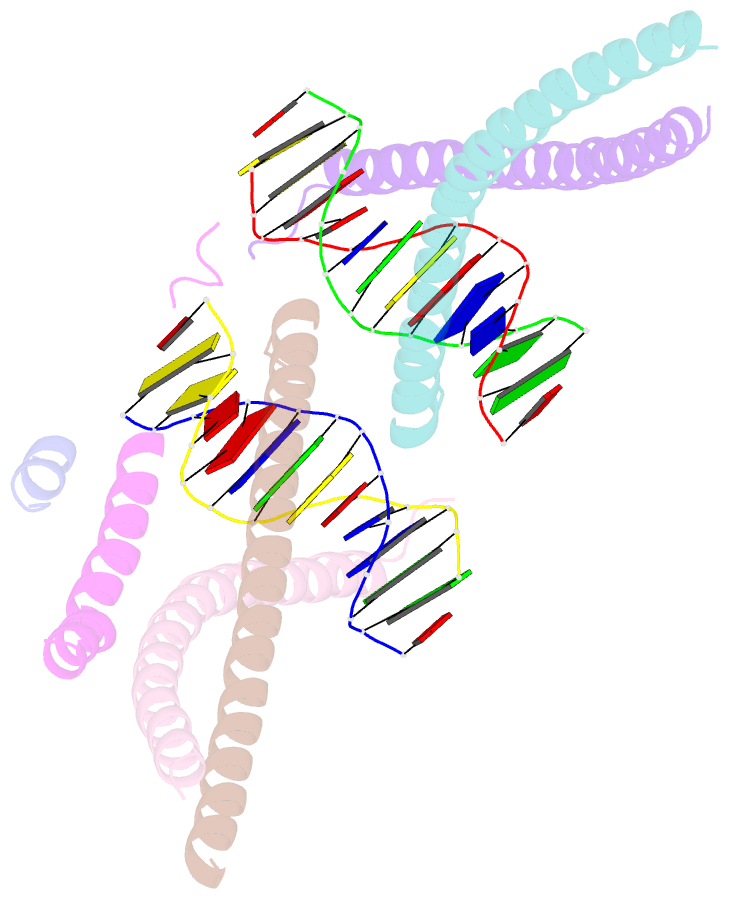
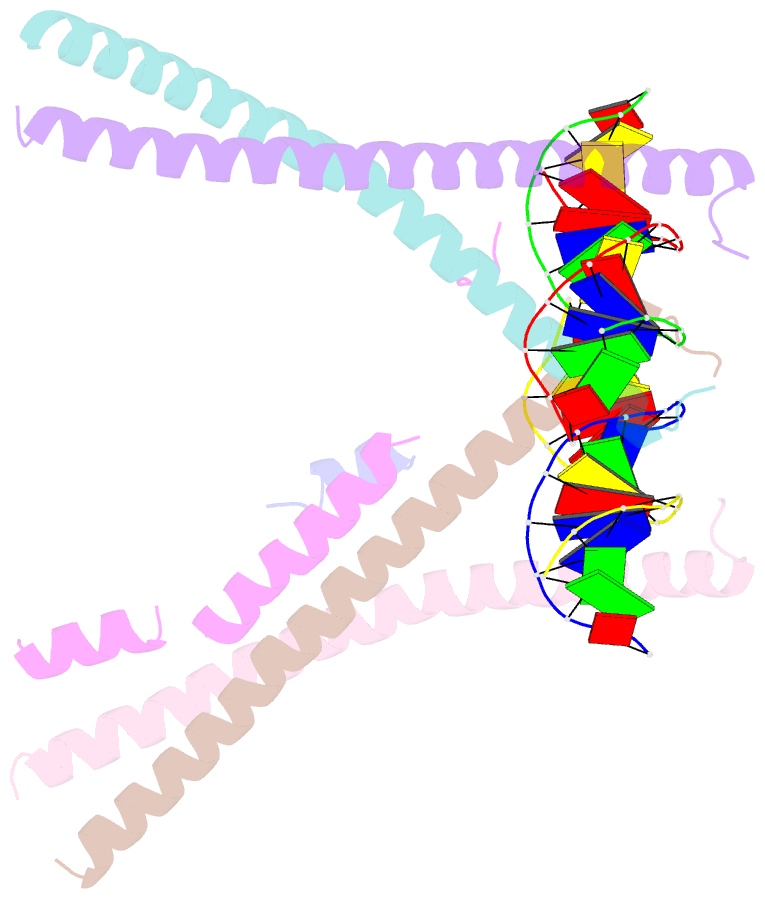
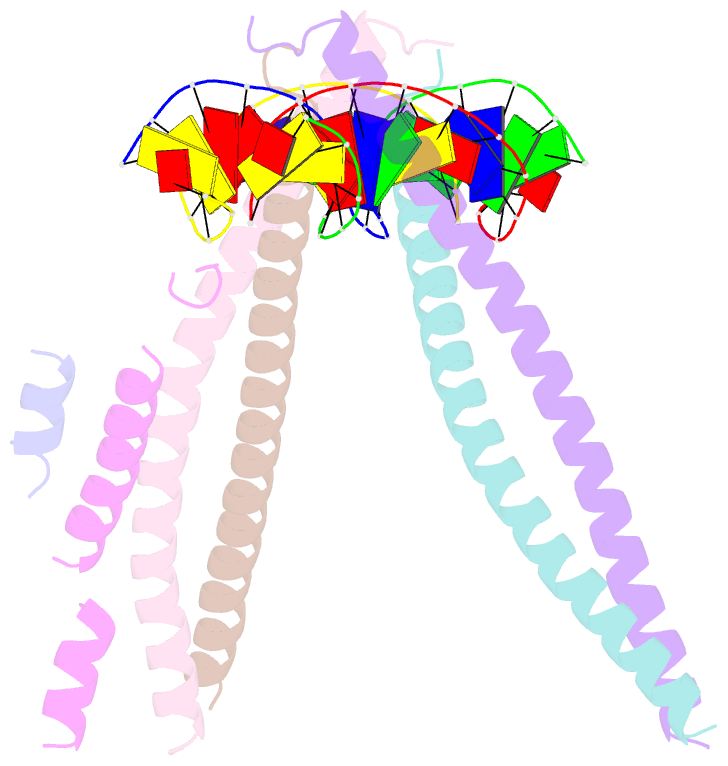
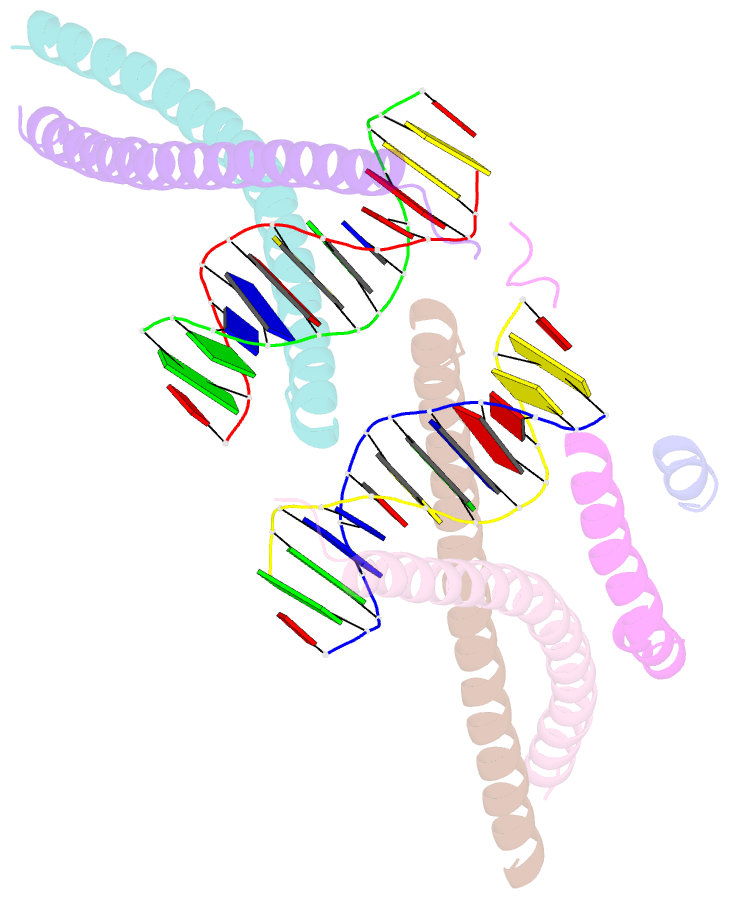
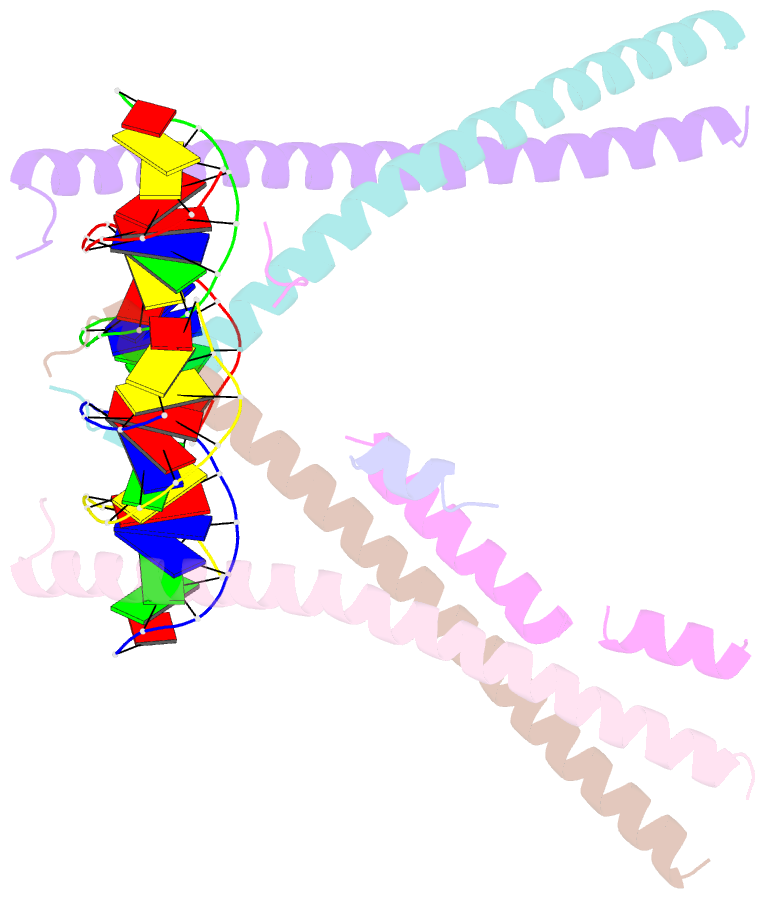
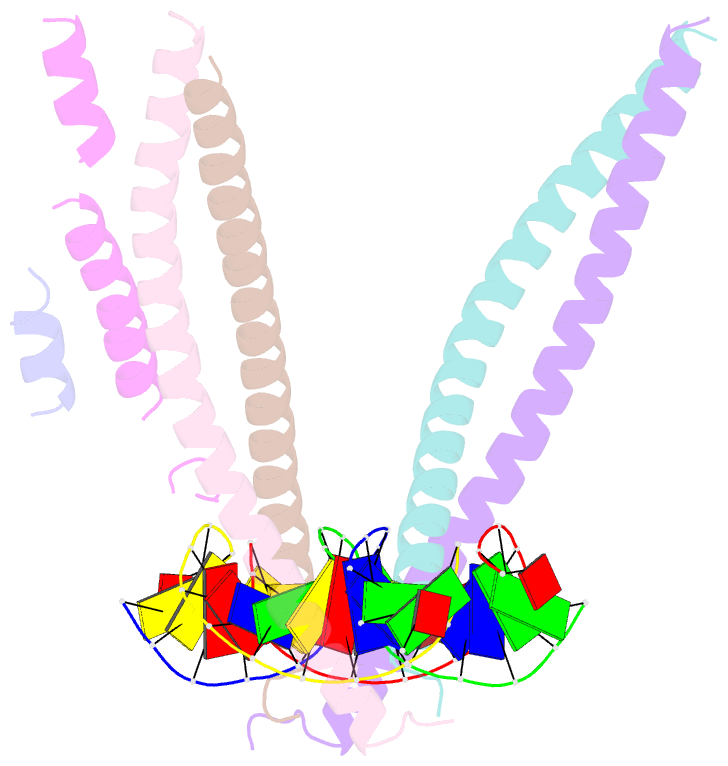
- Crystal structure of bzip transcription factor pap1 bound to DNA
- Reference
- Fujii Y, Shimizu T, Toda T, Yanagida M, Hakoshima T (2000): "Structural basis for the diversity of DNA recognition by bZIP transcription factors." Nat.Struct.Biol., 7, 889-893. doi: 10.1038/82822.
- Abstract
- The basic region leucine zipper (bZIP) proteins form one of the largest families of transcription factors in eukaryotic cells. Despite relatively high homology between the amino acid sequences of the bZIP motifs, these proteins recognize diverse DNA sequences. Here we report the 2.0 A resolution crystal structure of the bZIP motif of one such transcription factor, PAP1, a fission yeast AP-1-like transcription factor that binds DNA containing the novel consensus sequence TTACGTAA. The structure reveals how the Pap1-specific residues of the bZIP basic region recognize the target sequence and shows that the side chain of the invariant Asn in the bZIP motif adopts an alternative conformation in Pap1. This conformation, which is stabilized by a Pap1-specific residue and its associated water molecule, recognizes a different base in the target sequence from that in other bZIP subfamilies.