Summary information and primary citation
- PDB-id
- 1gtr; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- complex (ligase-trna)
- Method
- X-ray (2.5 Å)
- Summary
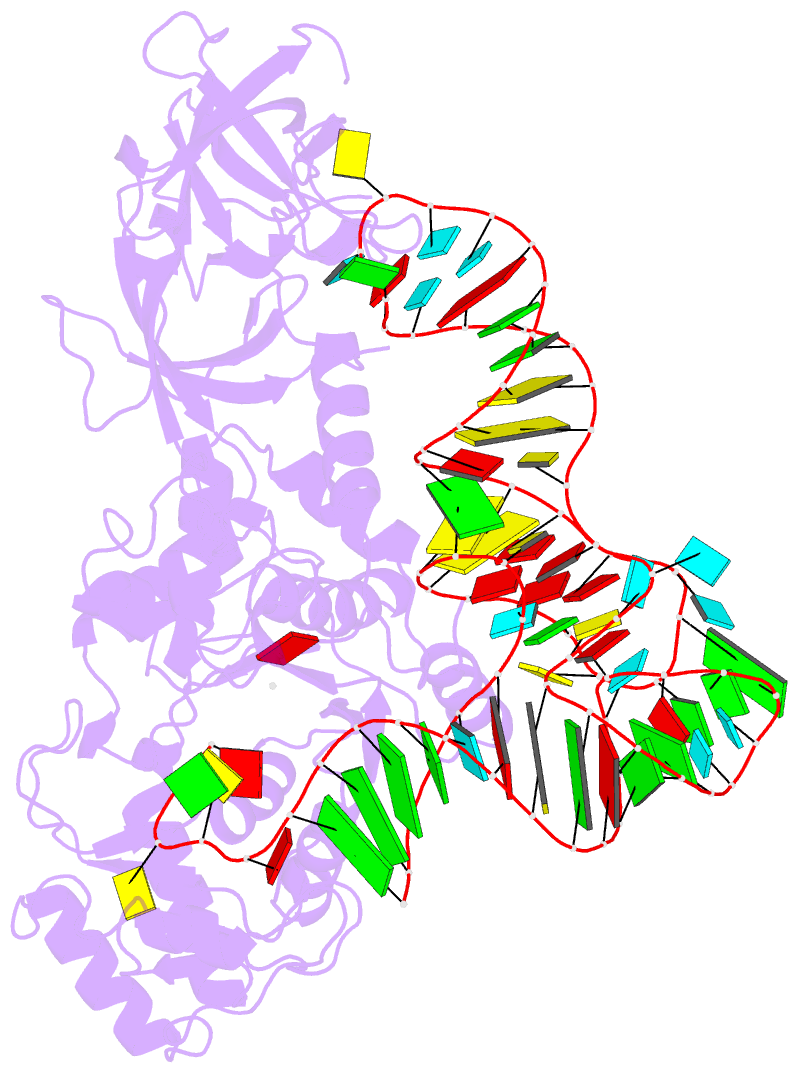
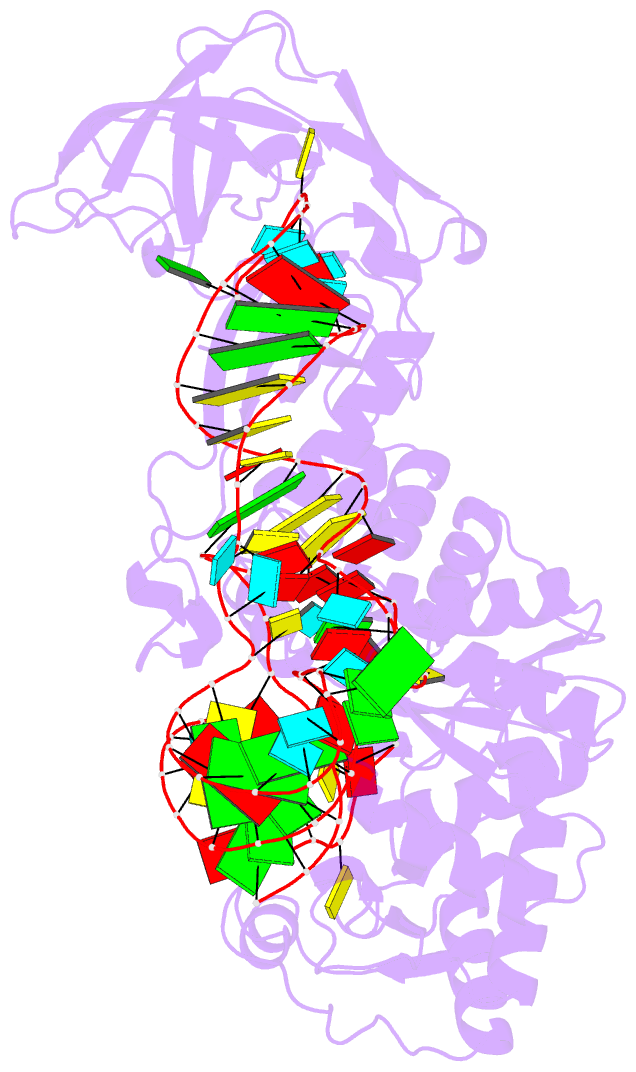
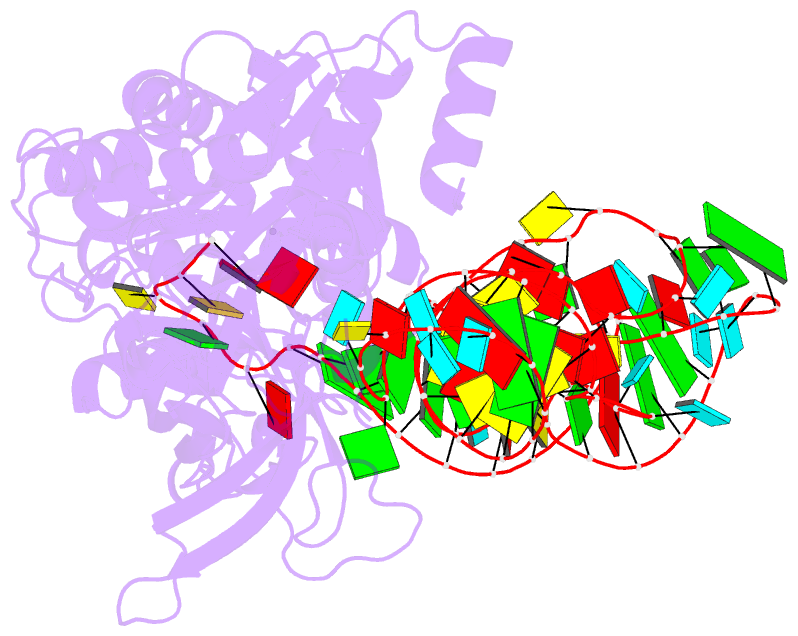
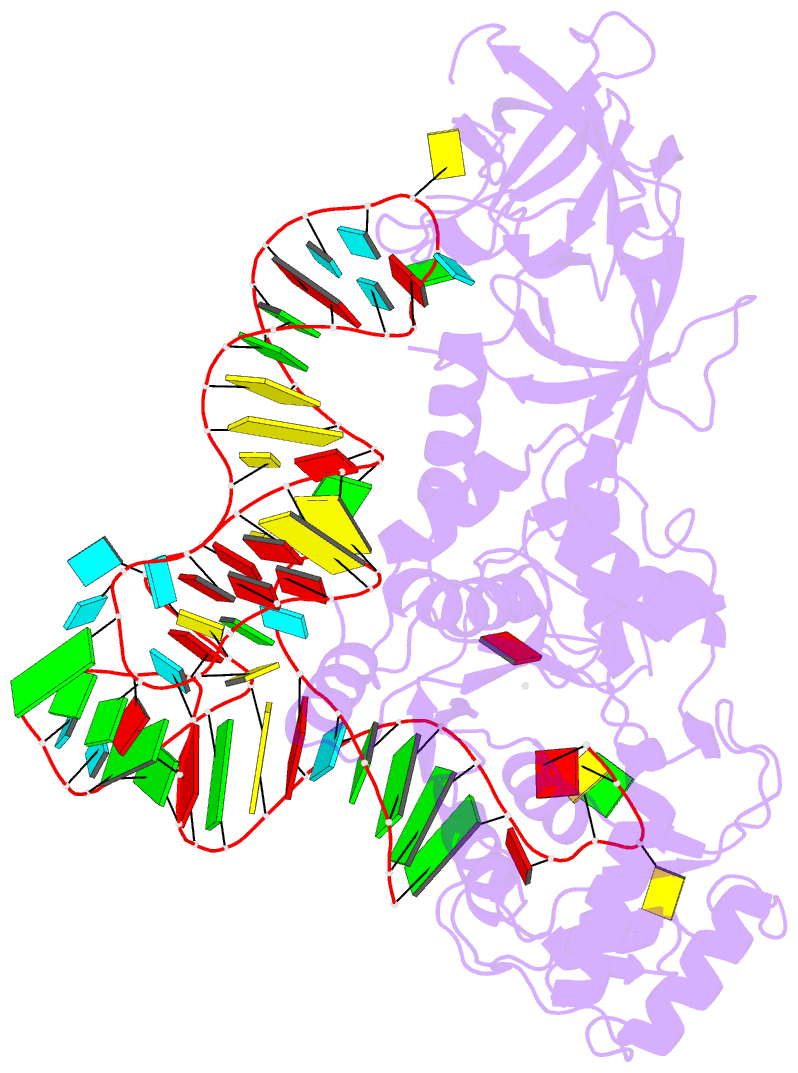
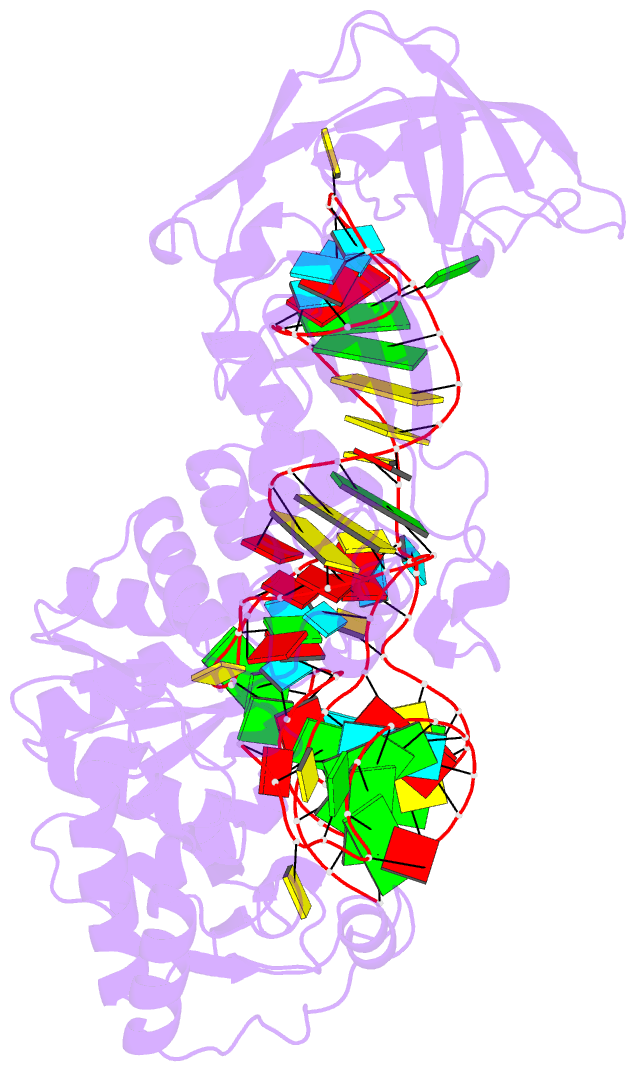
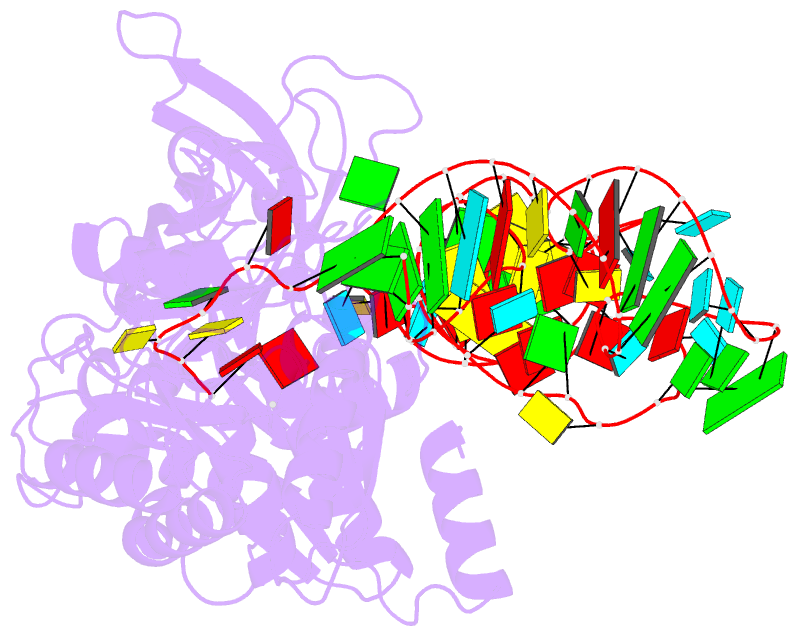
- Structural basis of anticodon loop recognition by glutaminyl-trna synthetase
- Reference
- Rould MA, Perona JJ, Steitz TA (1991): "Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase." Nature, 352, 213-218. doi: 10.1038/352213a0.
- Abstract
- The refined crystal structure of Escherichia coli glutaminyl transfer RNA synthetase complexed with transfer RNA(Gln) and ATP reveals that the structure of the anticodon loop of the enzyme-bound tRNA(Gln) differs extensively from that of the known crystal structures of uncomplexed tRNA molecules. The anticodon stem is extended by two non-Watson-Crick base pairs, leaving the three anti-codon bases unpaired and splayed out to bind snugly into three separate complementary pockets in the protein. These interactions suggest that the entire anticodon loop provides essential sites for glutaminyl tRNA synthetase discrimination among tRNA molecules.