Summary information and primary citation
- PDB-id
- 1h3e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase
- Method
- X-ray (2.9 Å)
- Summary
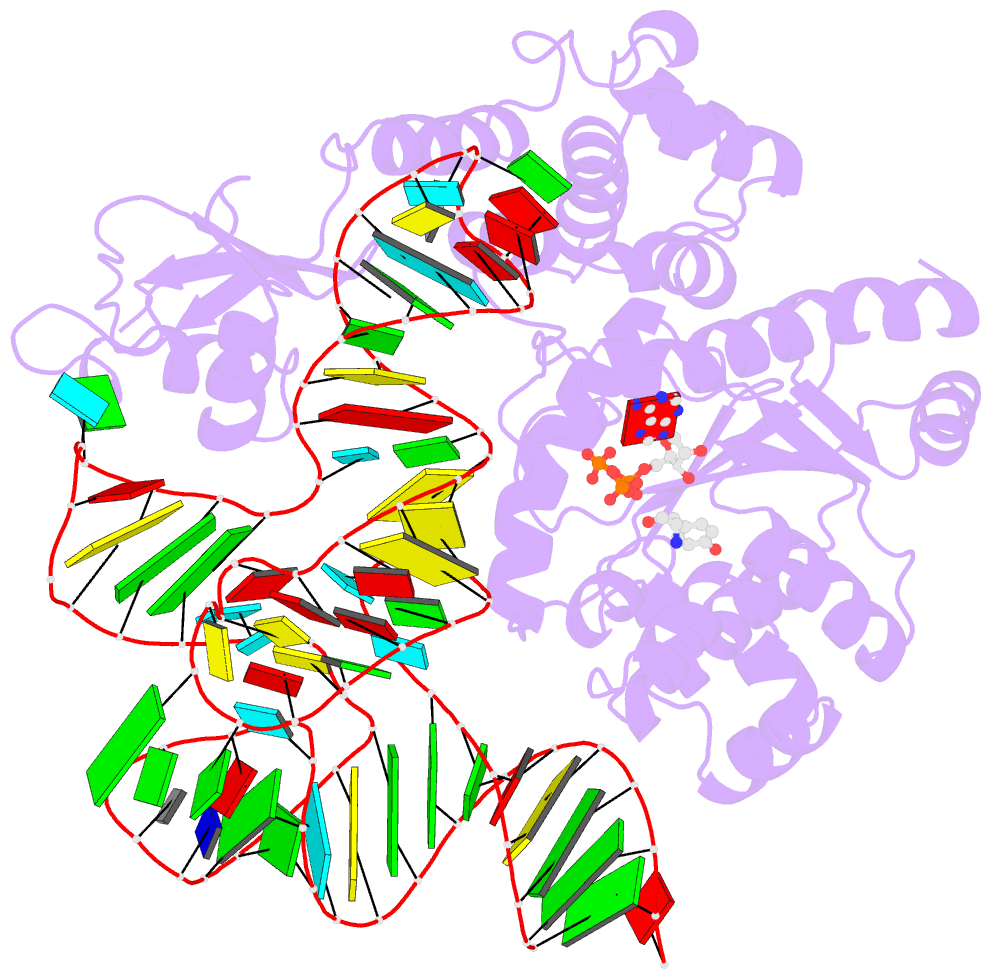
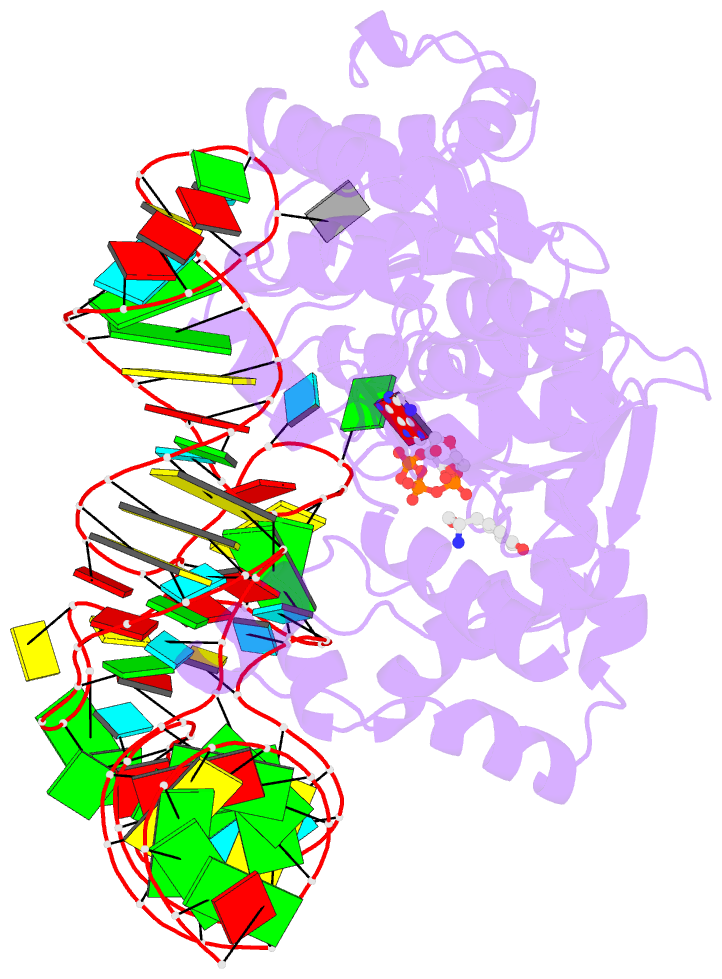
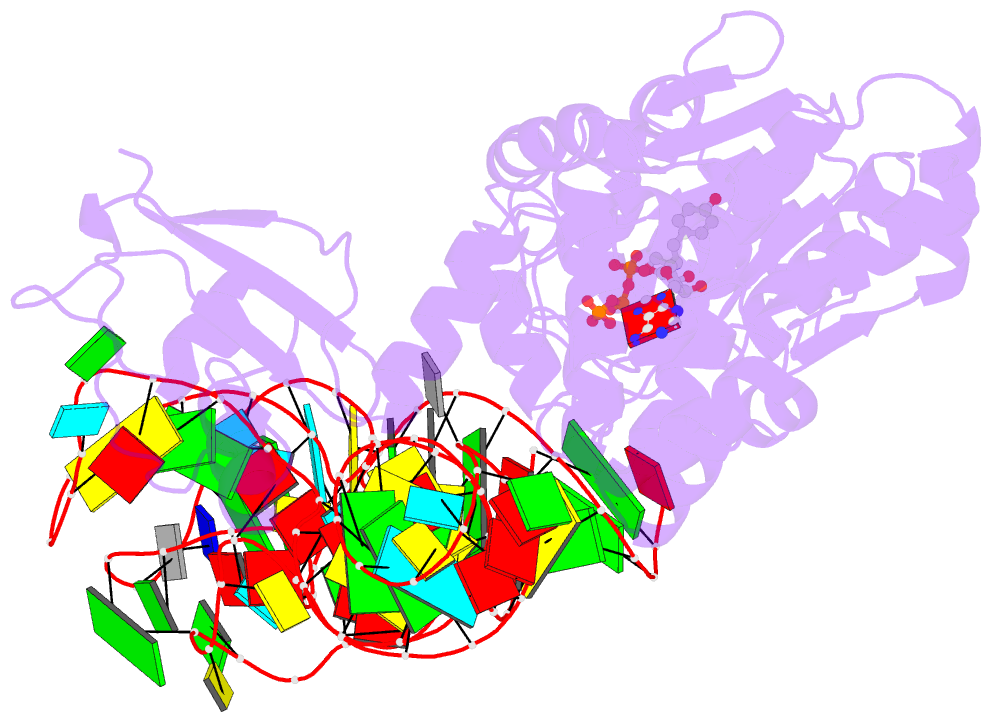
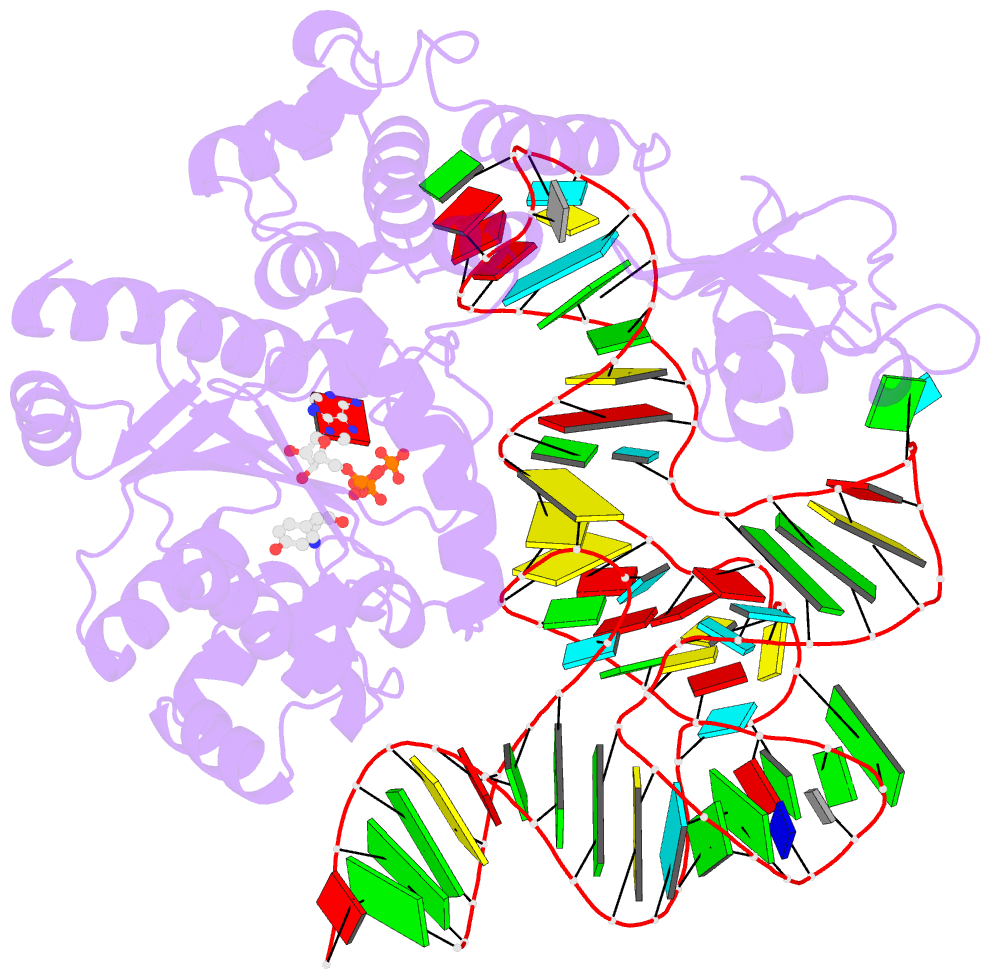
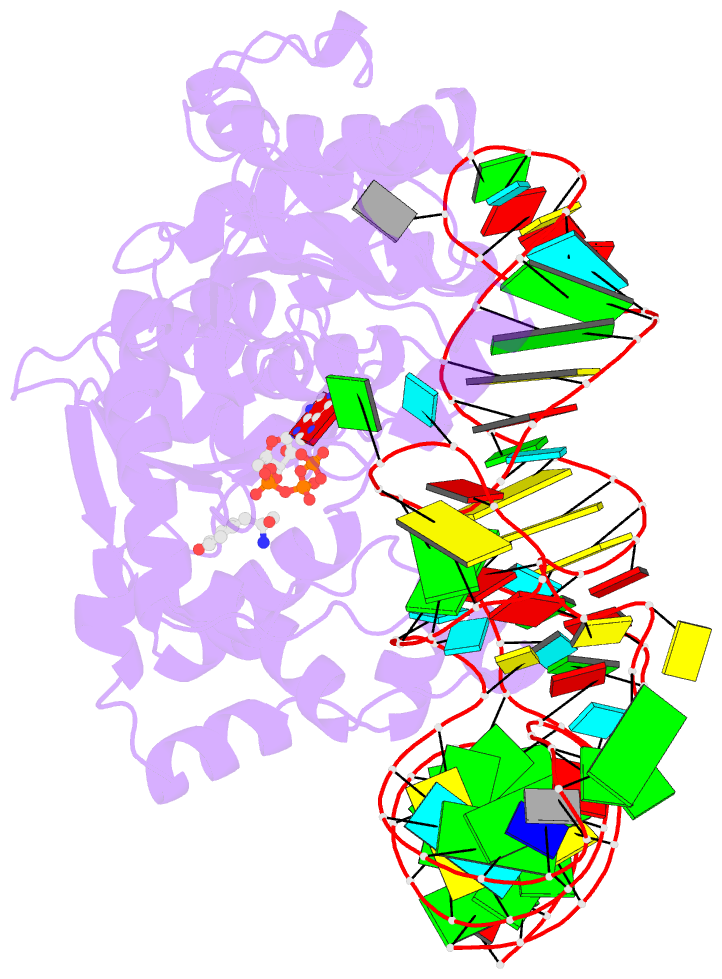
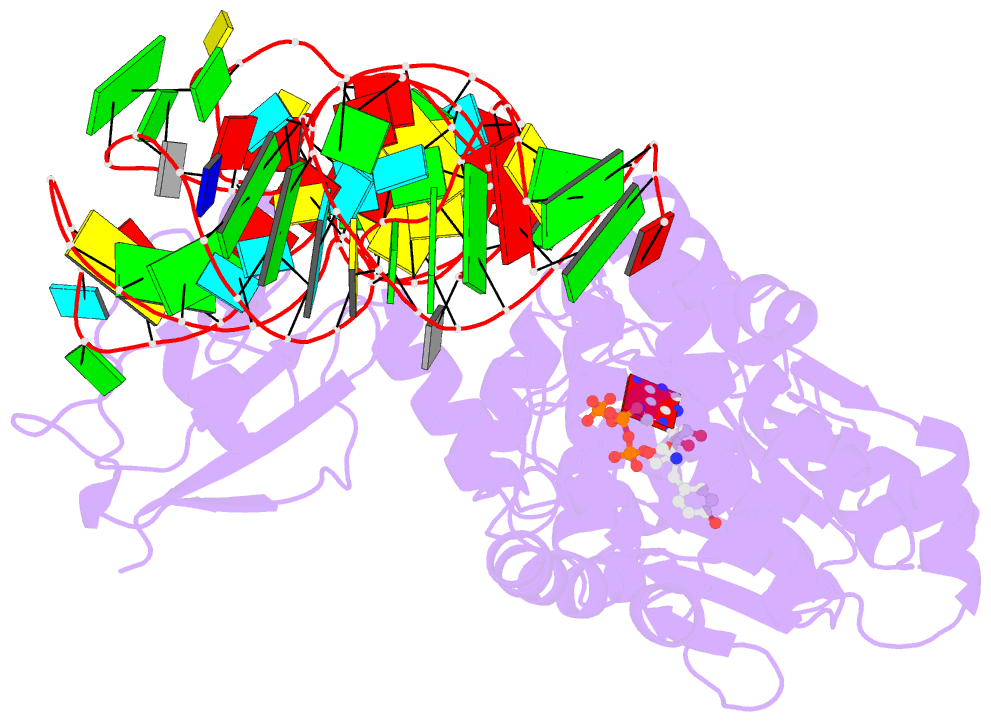
- Tyrosyl-trna synthetase from thermus thermophilus complexed with wild-type trnatyr(gua) and with atp and tyrosinol
- Reference
- Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S (2002): "Class I Tyrosyl-tRNA Synthetase Has a Class II Mode or tRNA Recognition." Embo J., 21, 3829. doi: 10.1093/EMBOJ/CDF373.
- Abstract
- Bacterial tyrosyl-tRNA synthetases (TyrRS) possess a flexibly linked C-terminal domain of approximately 80 residues, which has hitherto been disordered in crystal structures of the enzyme. We have determined the structure of Thermus thermophilus TyrRS at 2.0 A resolution in a crystal form in which the C-terminal domain is ordered, and confirm that the fold is similar to part of the C-terminal domain of ribosomal protein S4. We have also determined the structure at 2.9 A resolution of the complex of T.thermophilus TyrRS with cognate tRNA(tyr)(G Psi A). In this structure, the C-terminal domain binds between the characteristic long variable arm of the tRNA and the anti-codon stem, thus recognizing the unique shape of the tRNA. The anticodon bases have a novel conformation with A-36 stacked on G-34, and both G-34 and Psi-35 are base-specifically recognized. The tRNA binds across the two subunits of the dimeric enzyme and, remarkably, the mode of recognition of the class I TyrRS for its cognate tRNA resembles that of a class II synthetase in being from the major groove side of the acceptor stem.