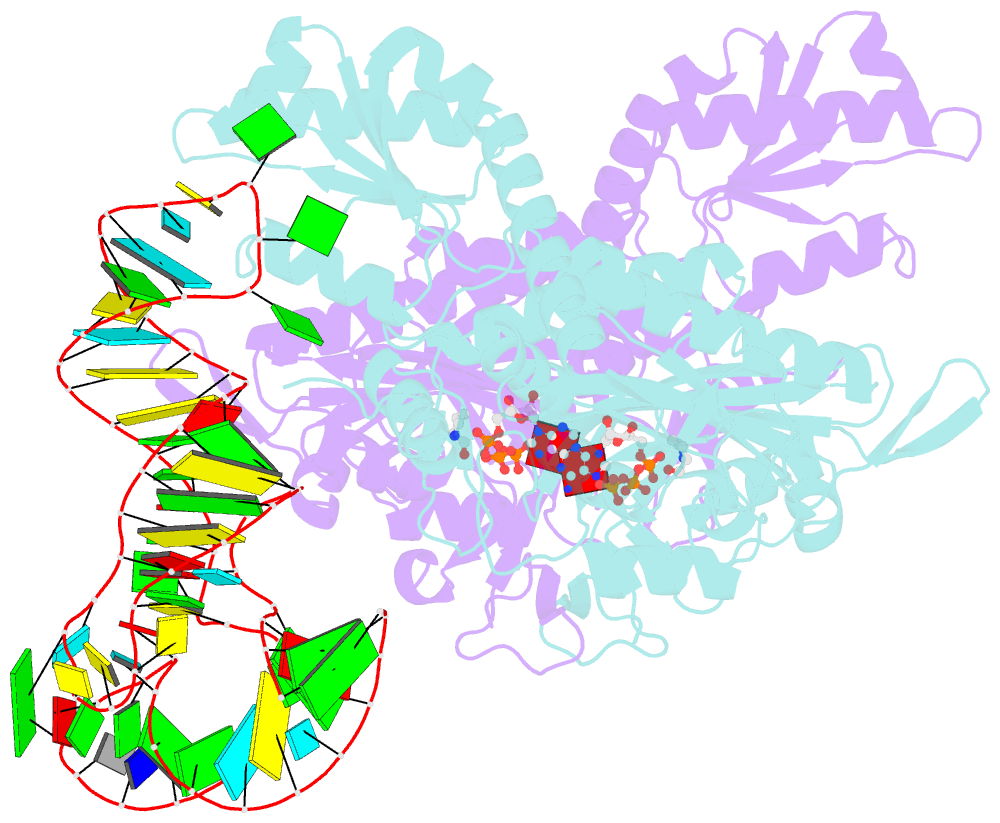
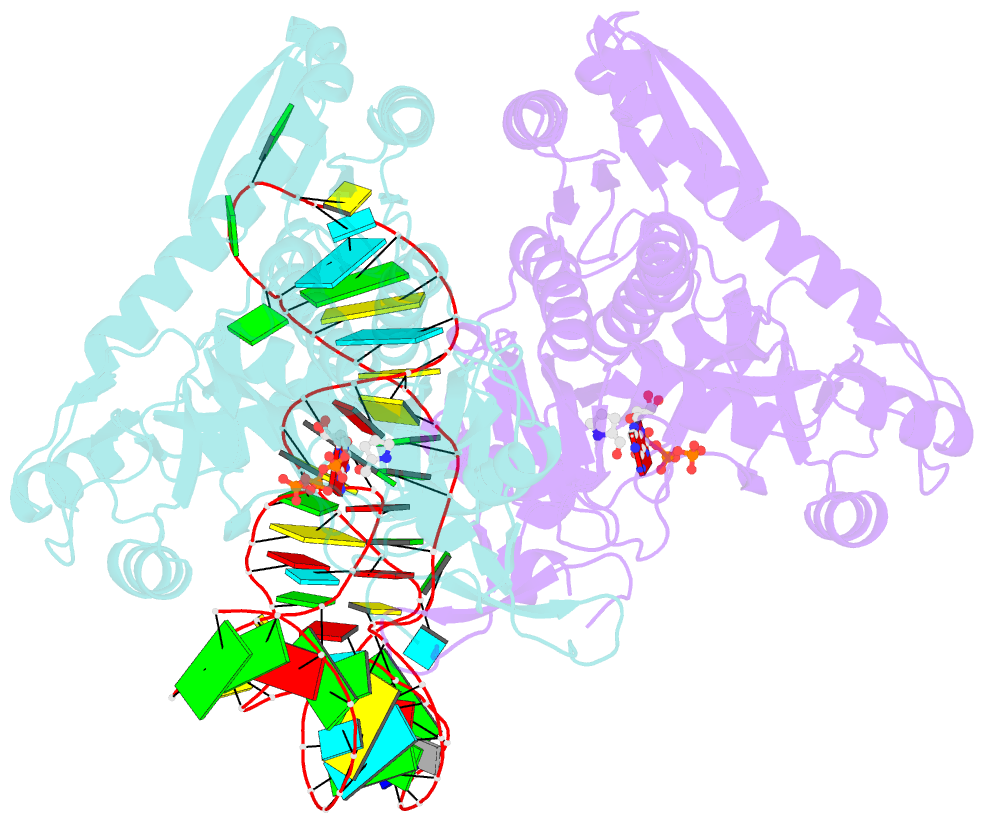
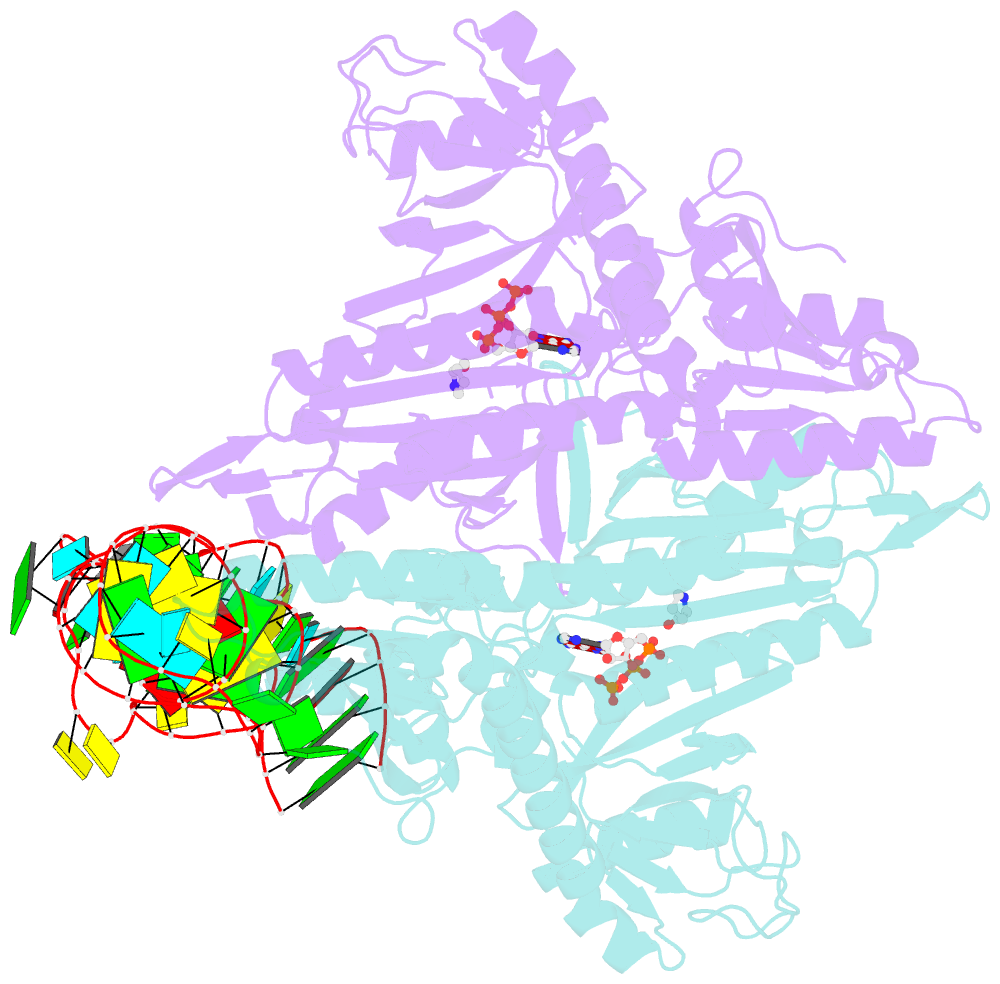
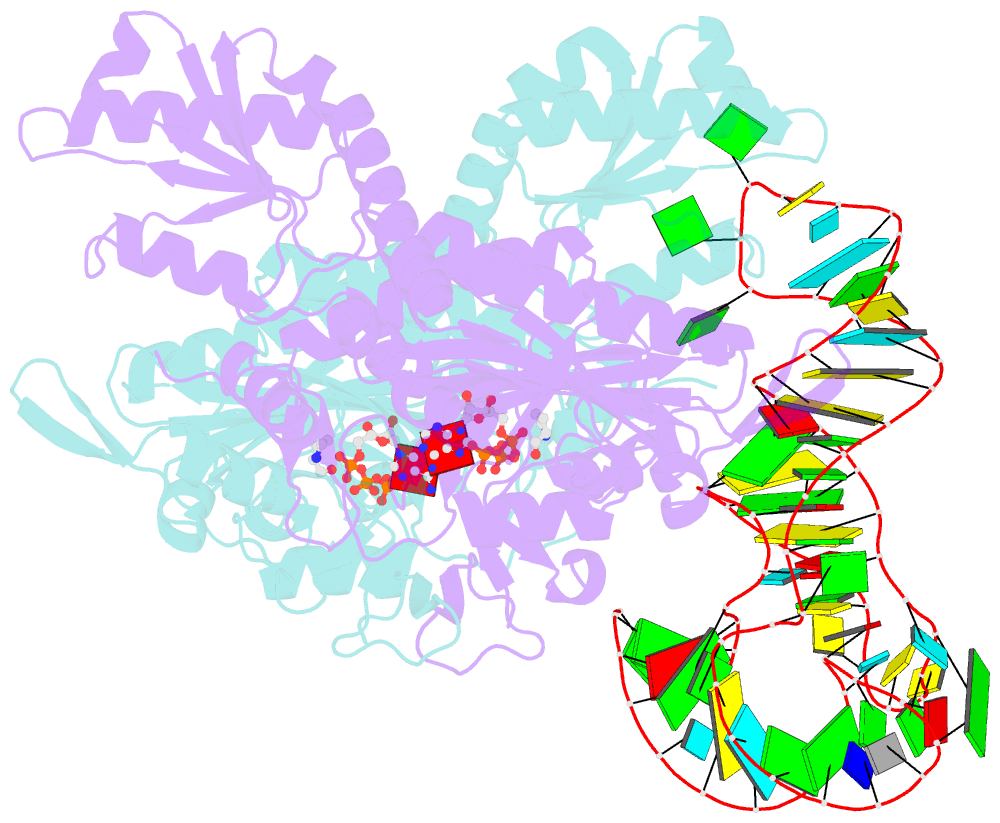
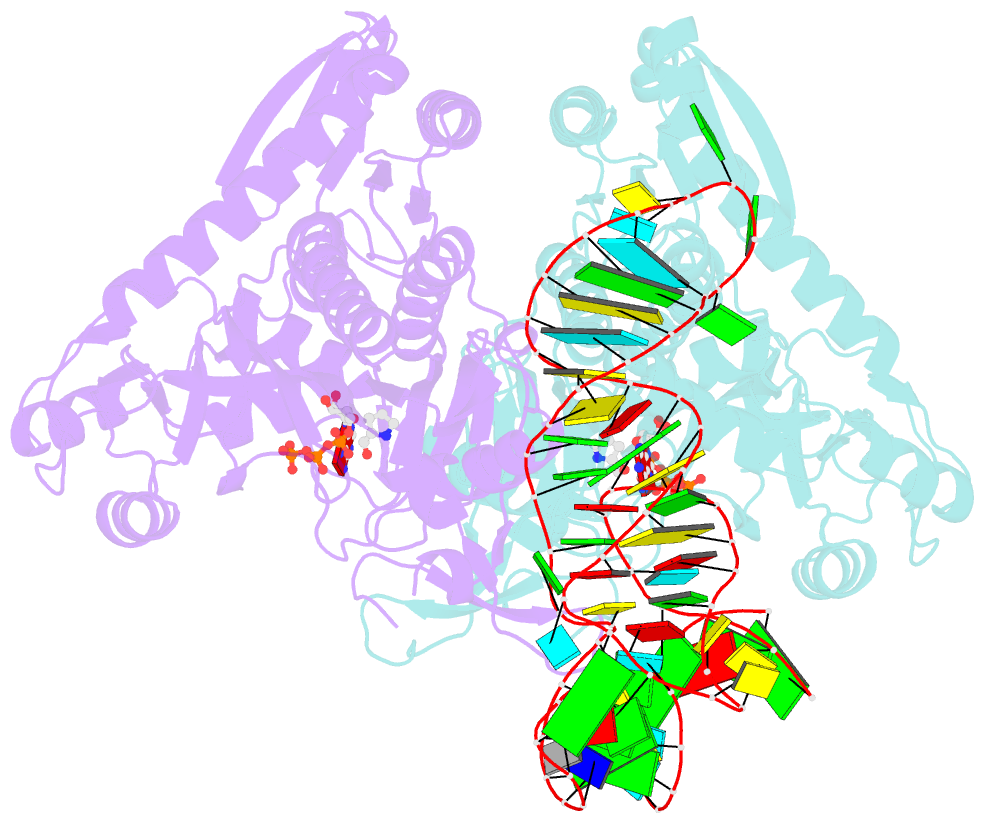
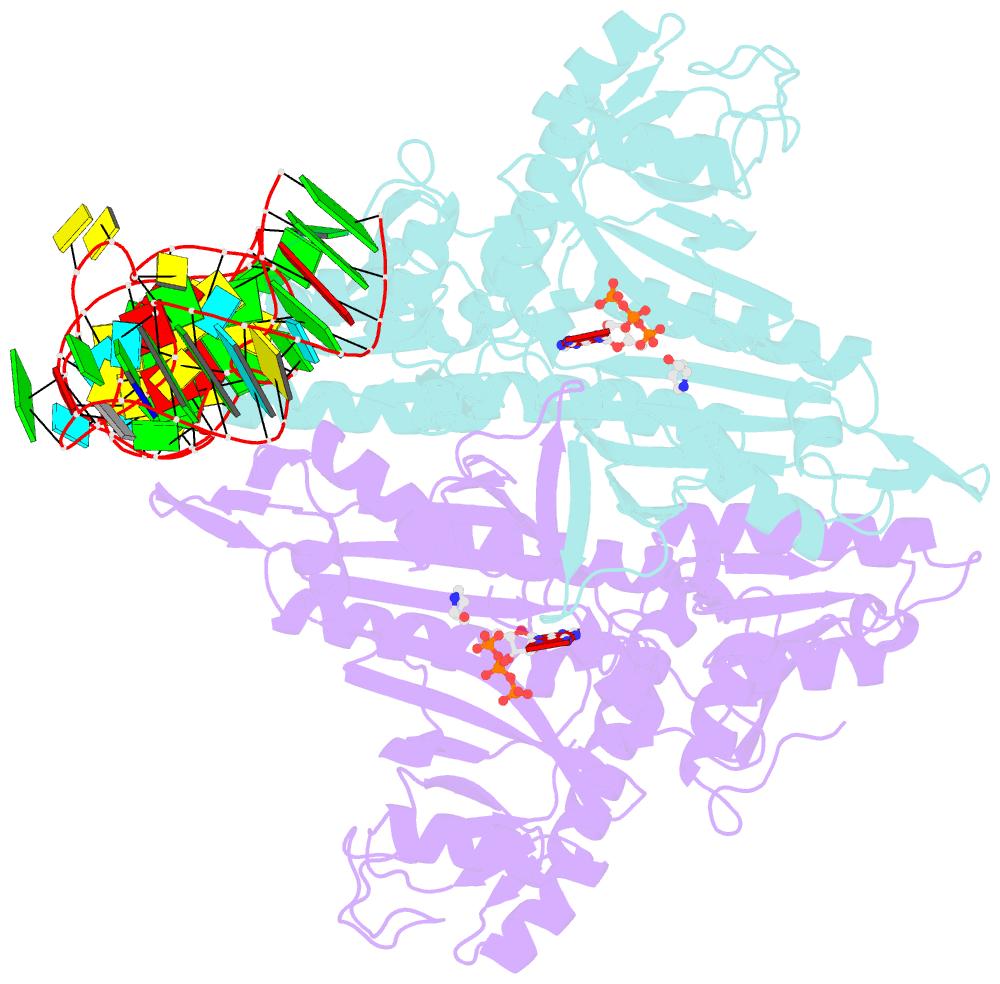
Summary information and primary citation
- PDB-id
- 1h4q; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- aminoacyl-trna synthetase
- Method
- X-ray (3.0 Å)
- Summary
- Prolyl-trna synthetase from thermus thermophilus complexed with trnapro(cgg), atp and prolinol
- Reference
- Yaremchuk A, Tukalo M, Grotli M, Cusack S (2001): "A Succession of Substrate Induced Conformational Changes Ensures the Amino Acid Specificity of Thermus Thermophilus Prolyl-tRNA Synthetase: Comparison with Histidyl-tRNA Synthetase." J.Mol.Biol., 309, 989. doi: 10.1006/JMBI.2001.4712.
- Abstract
- We describe the recognition by Thermus thermophilus prolyl-tRNA synthetase (ProRSTT) of proline, ATP and prolyl-adenylate and the sequential conformational changes occurring when the substrates bind and the activated intermediate is formed. Proline and ATP binding cause respectively conformational changes in the proline binding loop and motif 2 loop. However formation of the activated intermediate is necessary for the final conformational ordering of a ten residue peptide ("ordering loop") close to the active site which would appear to be essential for functional tRNA 3' end binding. These induced fit conformational changes ensure that the enzyme is highly specific for proline activation and aminoacylation. We also present new structures of apo and AMP bound histidyl-tRNA synthetase (HisRS) from T. thermophilus which we compare to our previous structures of the histidine and histidyl-adenylate bound enzyme. Qualitatively, similar results to those observed with T. thermophilus prolyl-tRNA synthetase are found. However histidine binding is sufficient to induce the co-operative ordering of the topologically equivalent histidine binding loop and ordering loop. These two examples contrast with most other class II aminoacyl-tRNA synthetases whose pocket for the cognate amino acid side-chain is largely preformed. T. thermophilus prolyl-tRNA synthetase appears to be the second class II aminoacyl-tRNA synthetase, after HisRS, to use a positively charged amino acid instead of a divalent cation to catalyse the amino acid activation reaction.