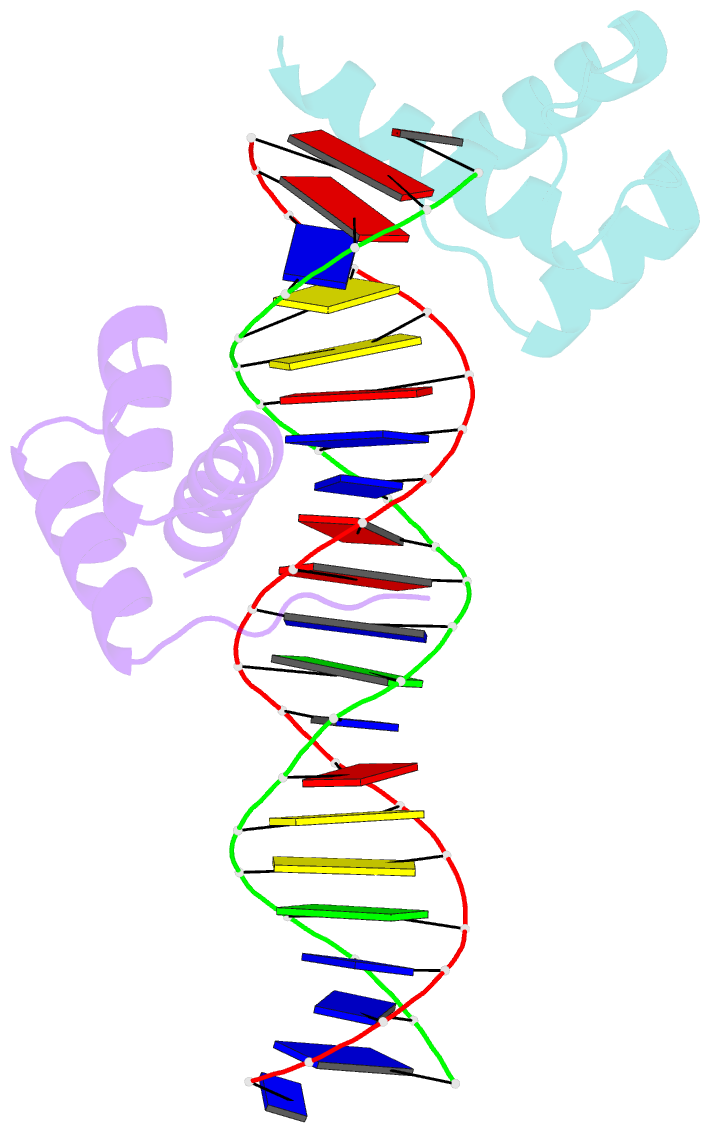
Summary information and primary citation
- PDB-id
- 1hdd; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.8 Å)
- Summary
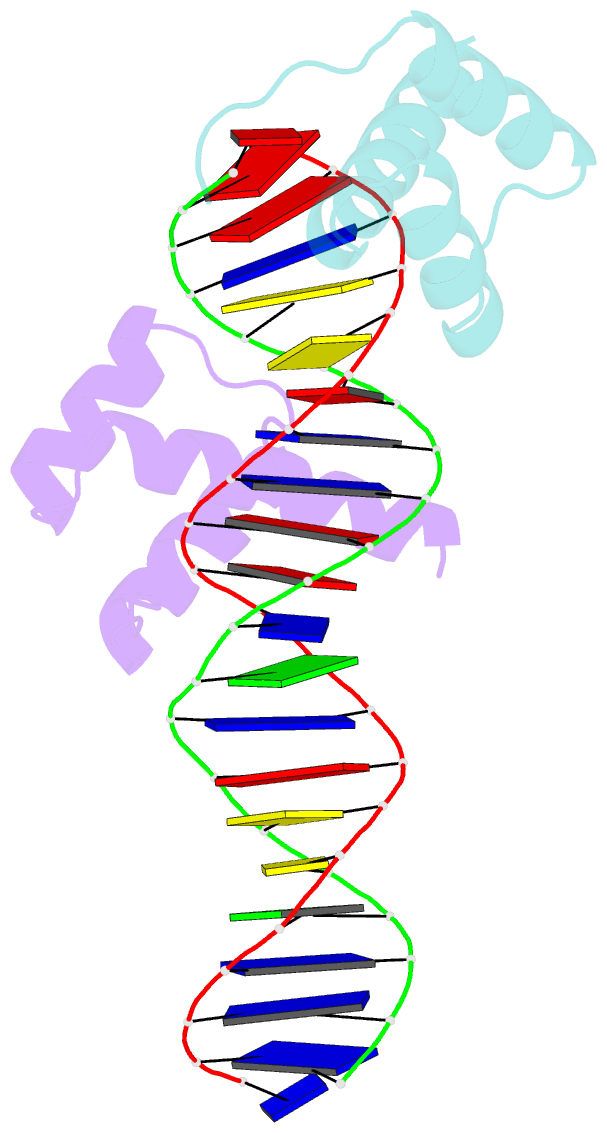
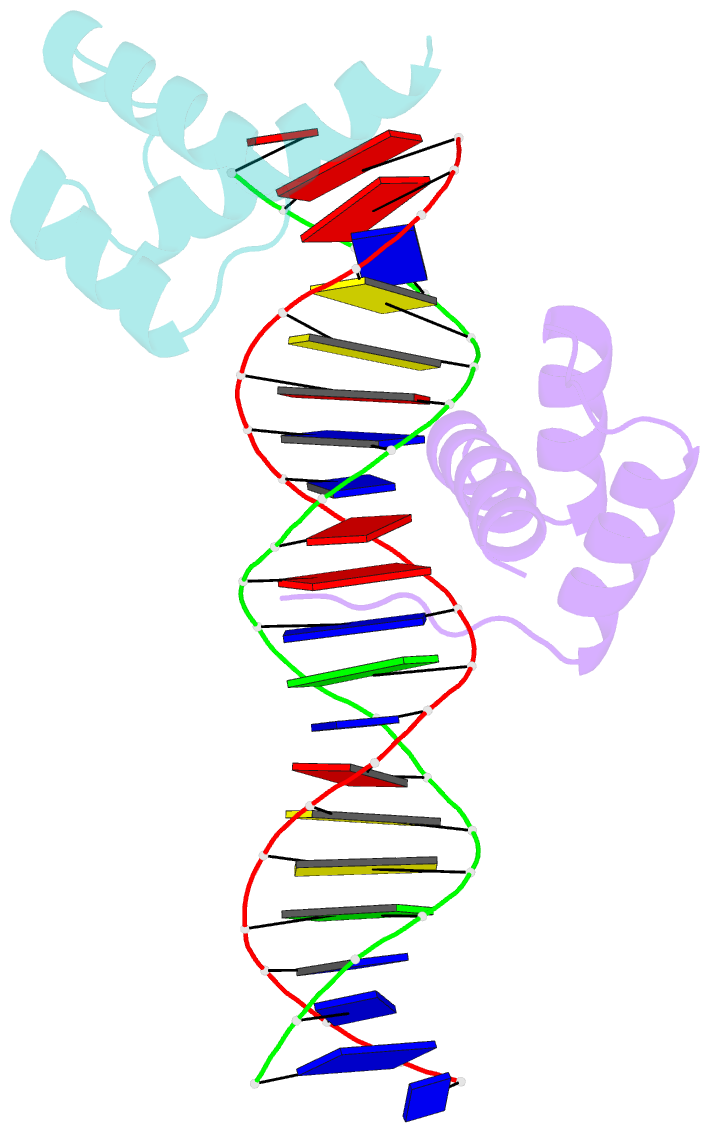
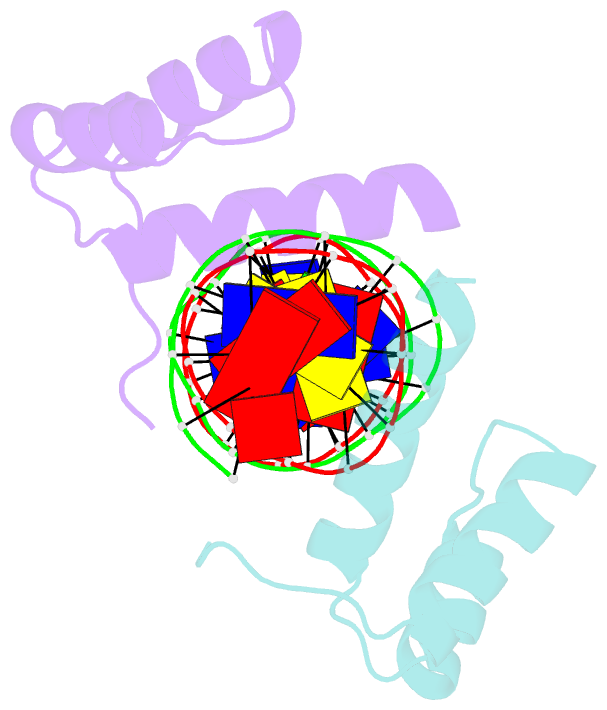
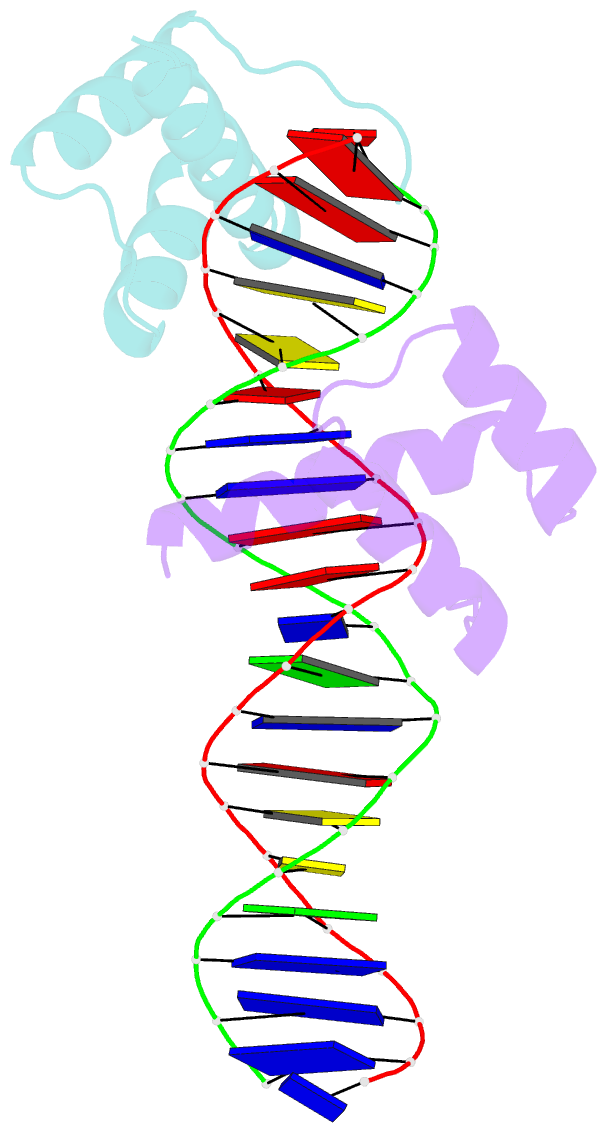
- Crystal structure of an engrailed homeodomain-DNA complex at 2.8 angstroms resolution: a framework for understanding homeodomain-DNA interactions
- Reference
- Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO (1990): "Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions." Cell(Cambridge,Mass.), 63, 579-590. doi: 10.1016/0092-8674(90)90453-L.
- Abstract
- The crystal structure of a complex containing the engrailed homeodomain and a duplex DNA site has been determined at 2.8 A resolution and refined to a crystallographic R factor of 24.4%. In this complex, two separate regions of the 61 amino acid polypeptide contact a TAAT subsite. An N-terminal arm fits into the minor groove, and the side chains of Arg-3 and Arg-5 make contacts near the 5' end of this "core consensus" binding site. An alpha helix fits into the major groove, and the side chains of IIe-47 and Asn-51 contact base pairs near the 3' end of the TAAT site. This "recognition helix" is part of a structurally conserved helix-turn-helix unit, but these helices are longer than the corresponding helices in the lambda repressor, and the relationship between the helix-turn-helix unit and the DNA is significantly different.