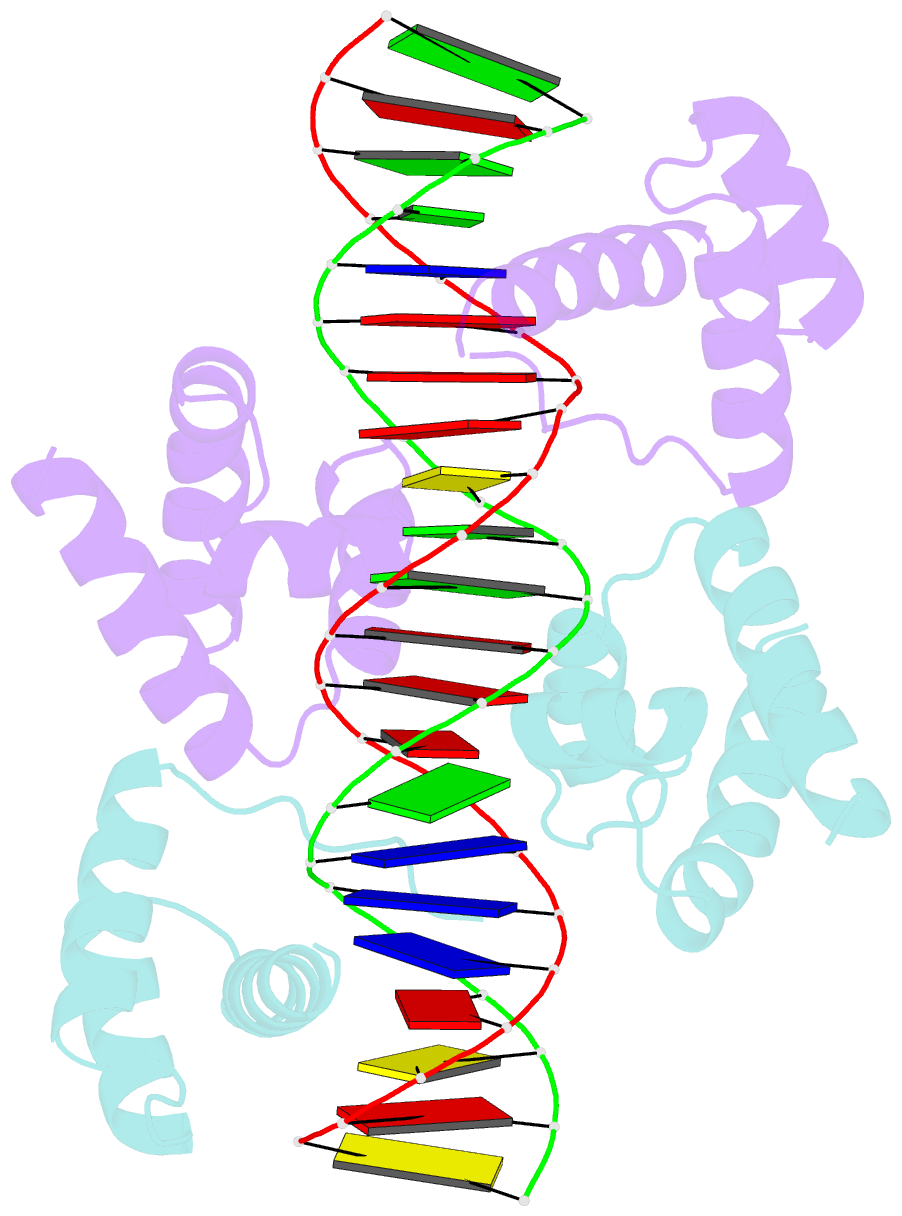
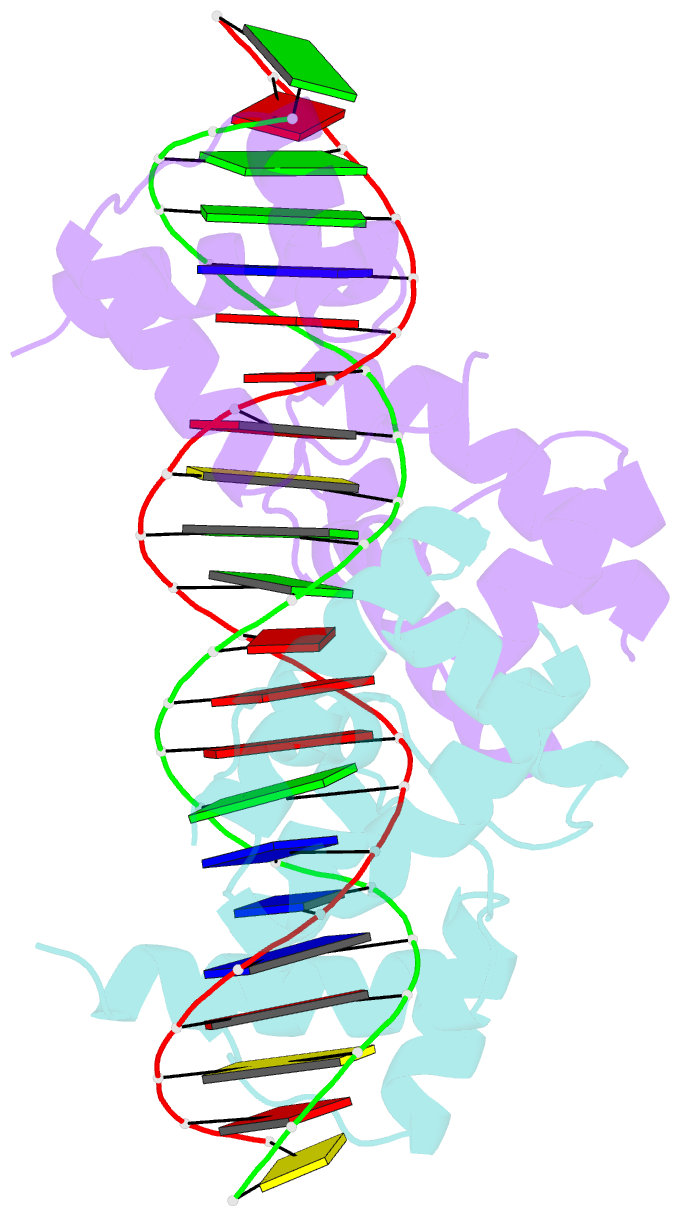
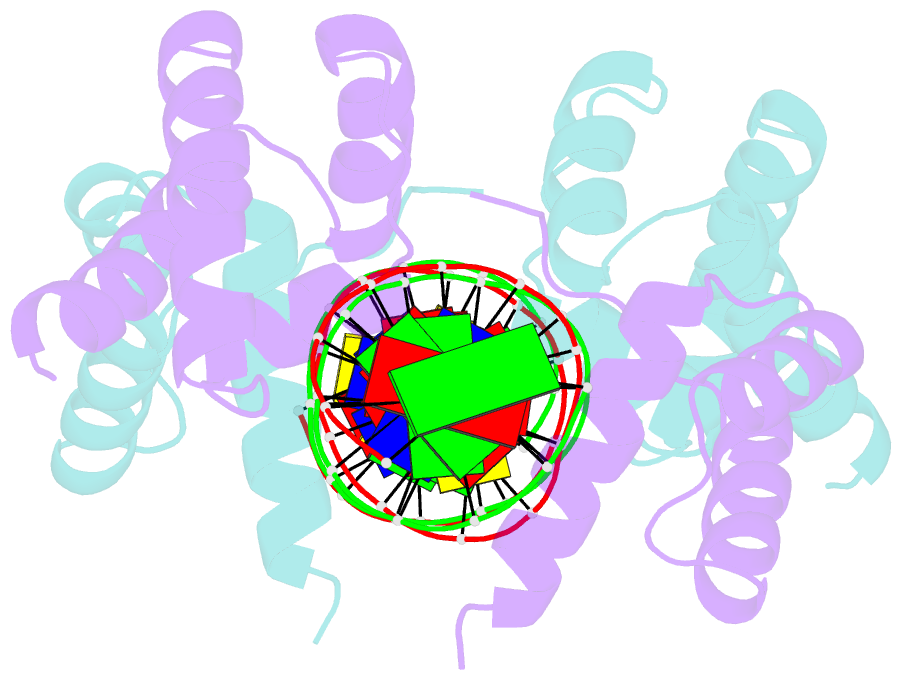
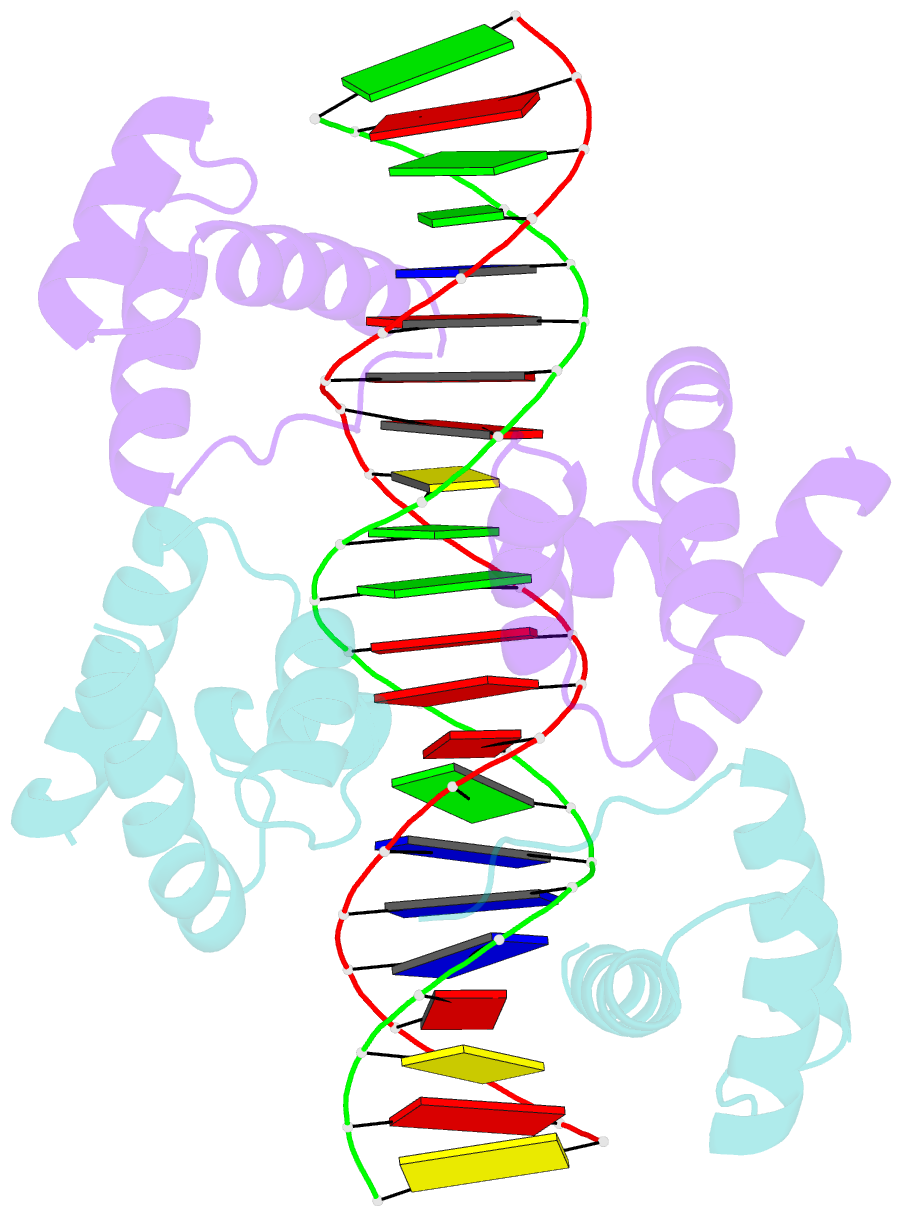
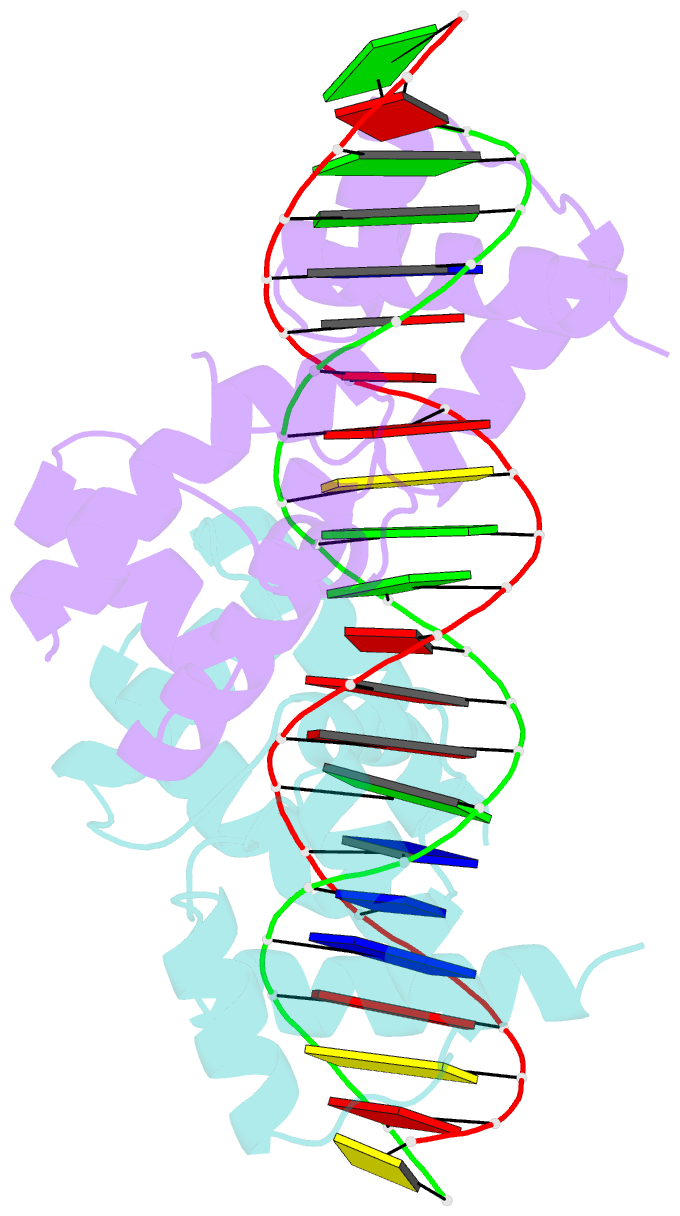
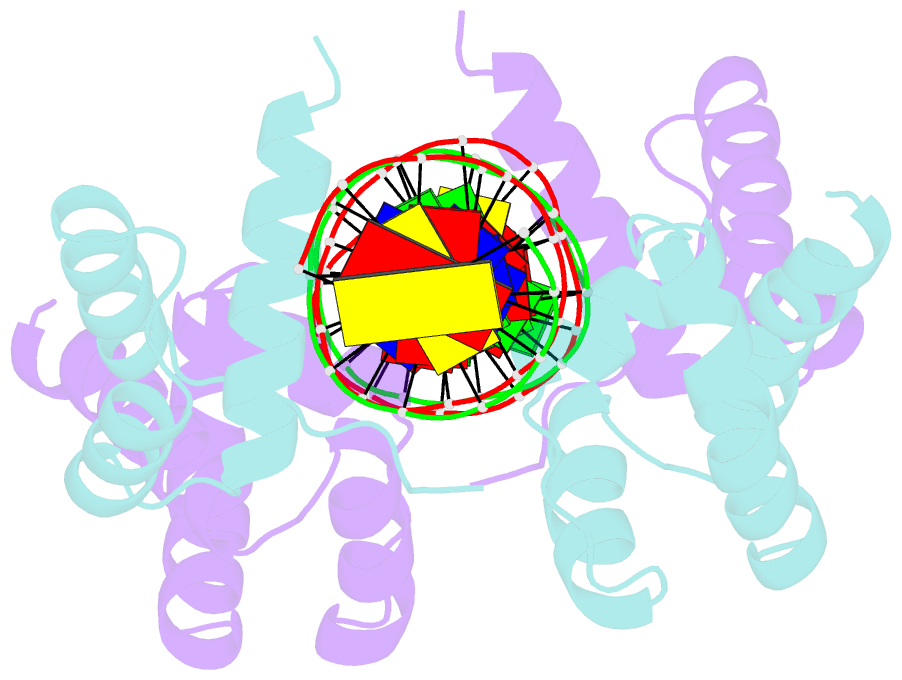
Summary information and primary citation
- PDB-id
- 1hf0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (2.7 Å)
- Summary
- Crystal structure of the DNA-binding domain of oct-1 bound to DNA as a dimer
- Reference
- Remenyi A, Tomilin A, Pohl E, Lins K, Philippsen A, Reinbold R, Scholer HR, Wilmanns M (2001): "Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping." Mol.Cell, 8, 569. doi: 10.1016/S1097-2765(01)00336-7.
- Abstract
- Two crystal structures of Oct-1 POU domain bound to DNA provide a rationale for differential, conformation-dependent recruitment of transcription cofactors. The POU-homeo and POU-specific subdomains of Oct-1 contain two different nonoverlapping pairs of surface patches that are capable of forming unrelated protein-protein interfaces. Members of the POU factor family contain one or two conserved sequence motifs in the interface that are known to be phosphorylated, as noted for Oct-1 and Pit-1. Modeling of Oct-4 reveals the unique case where the same conserved sequence is located in both interfaces. Our studies provide the basis for two distinct dimeric POU factor arrangements that are dictated by the architecture of each DNA response element. We suggest interface swapping in dimers could be a general mechanism of modulating the activity of transcription factors.