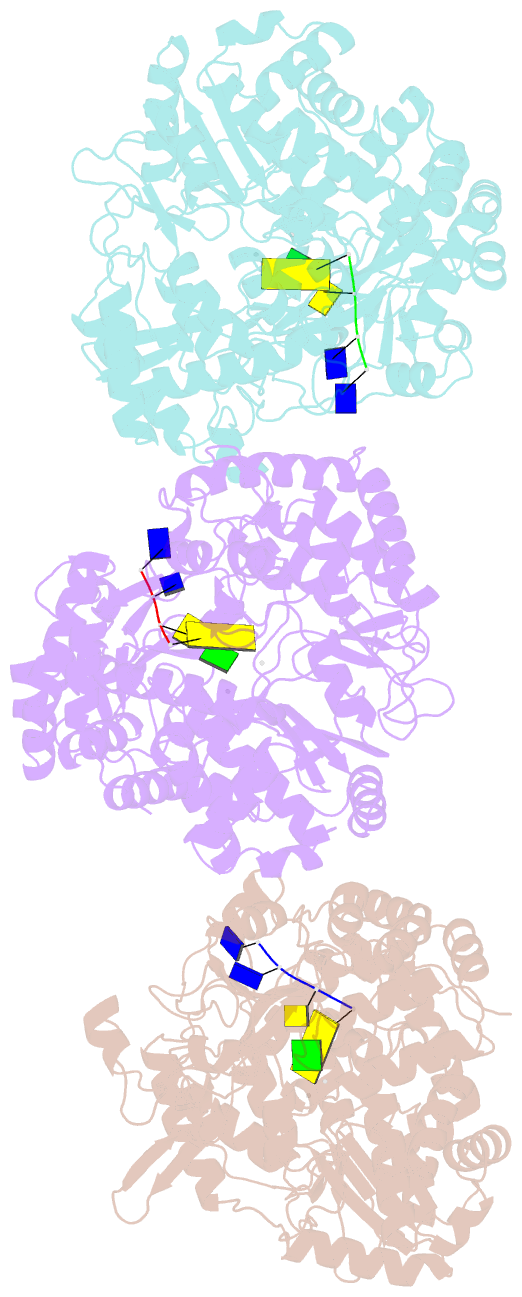
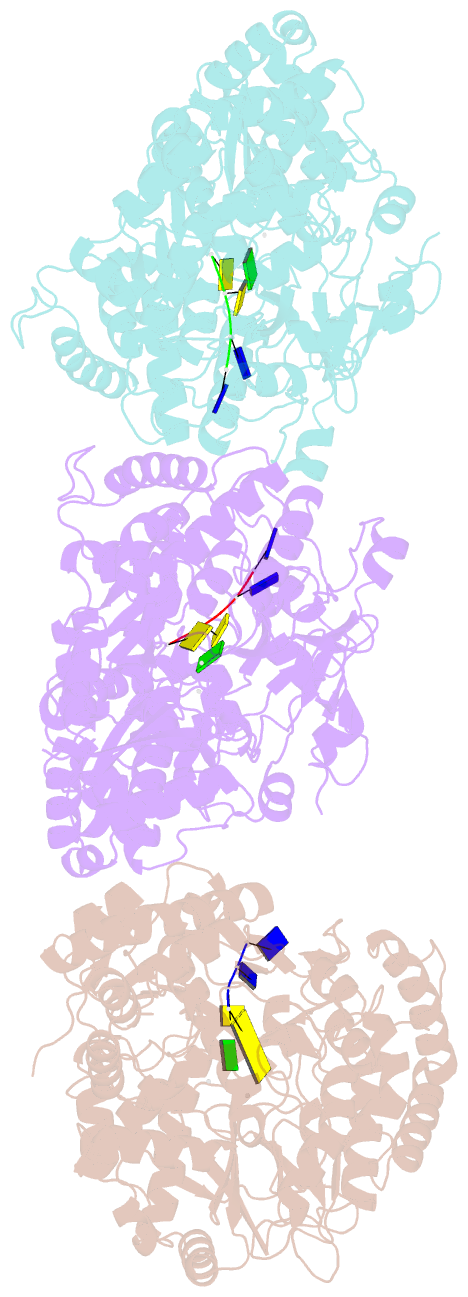
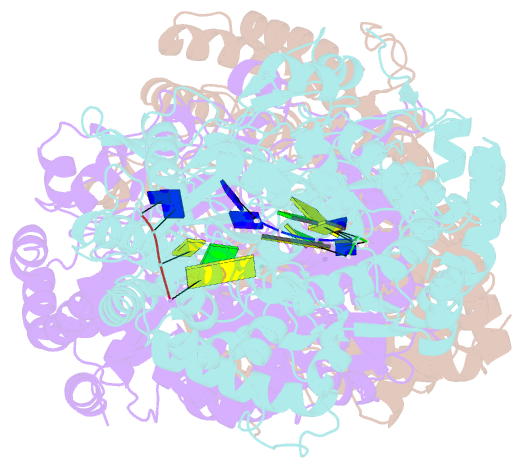
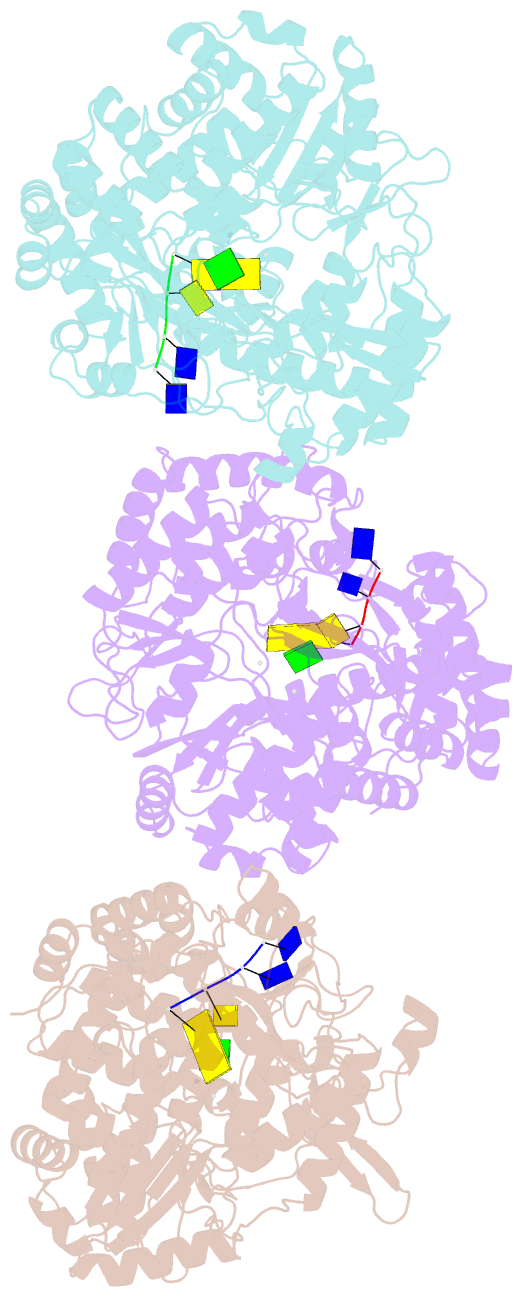
Summary information and primary citation
- PDB-id
- 1hi0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA polymerase
- Method
- X-ray (3.0 Å)
- Summary
- RNA dependent RNA polymerase from dsrna bacteriophage phi6 plus initiation complex
- Reference
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI (2001): "A Mechanism for Initiating RNA-Dependent RNA Polymerization." Nature, 410, 235. doi: 10.1038/35065653.
- Abstract
- In most RNA viruses, genome replication and transcription are catalysed by a viral RNA-dependent RNA polymerase. Double-stranded RNA viruses perform these operations in a capsid (the polymerase complex), using an enzyme that can read both single- and double-stranded RNA. Structures have been solved for such viral capsids, but they do not resolve the polymerase subunits in any detail. Here we show that the 2 A resolution X-ray structure of the active polymerase subunit from the double-stranded RNA bacteriophage straight phi6 is highly similar to that of the polymerase of hepatitis C virus, providing an evolutionary link between double-stranded RNA viruses and flaviviruses. By crystal soaking and co-crystallization, we determined a number of other structures, including complexes with oligonucleotide and/or nucleoside triphosphates (NTPs), that suggest a mechanism by which the incoming double-stranded RNA is opened up to feed the template through to the active site, while the substrates enter by another route. The template strand initially overshoots, locking into a specificity pocket, and then, in the presence of cognate NTPs, reverses to form the initiation complex; this process engages two NTPs, one of which acts with the carboxy-terminal domain of the protein to prime the reaction. Our results provide a working model for the initiation of replication and transcription.