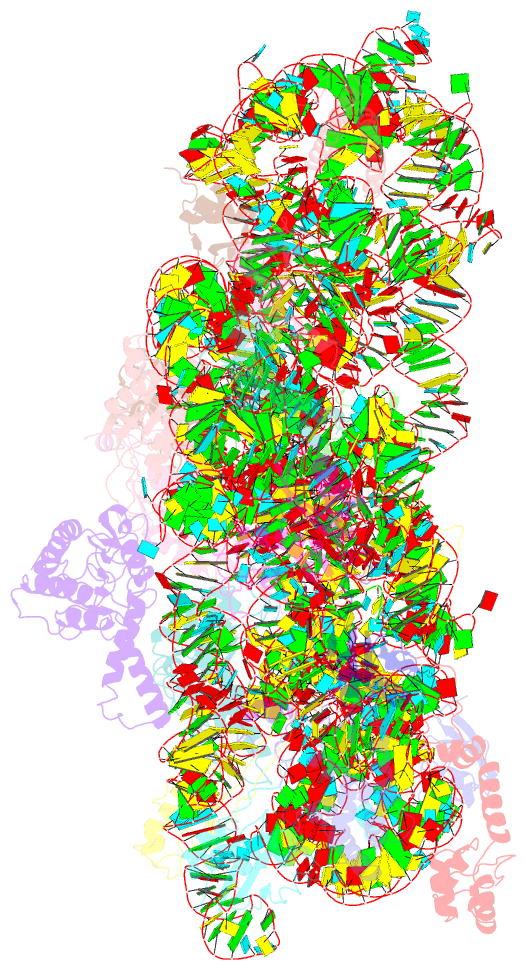
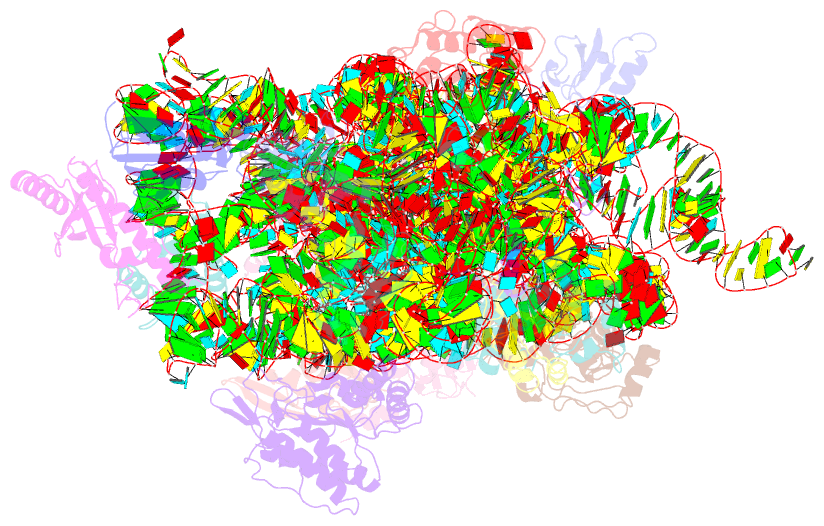
Summary information and primary citation
- PDB-id
-
1hr0;
DSSR-derived features in text and
JSON formats; DNAproDB
- Class
- ribosome
- Method
- X-ray (3.2 Å)
- Summary
- Crystal structure of initiation factor if1 bound to the
30s ribosomal subunit
- Reference
-
Carter AP, Clemons Jr WM, Brodersen DE, Morgan-Warren RJ,
Hartsch T, Wimberly BT, Ramakrishnan V (2001): "Crystal
structure of an initiation factor bound to the 30S
ribosomal subunit." Science,
291, 498-501. doi: 10.1126/science.1057766.
- Abstract
- Initiation of translation at the correct position on
messenger RNA is essential for accurate protein synthesis.
In prokaryotes, this process requires three initiation
factors: IF1, IF2, and IF3. Here we report the crystal
structure of a complex of IF1 and the 30S ribosomal
subunit. Binding of IF1 occludes the ribosomal A site and
flips out the functionally important bases A1492 and A1493
from helix 44 of 16S RNA, burying them in pockets in IF1.
The binding of IF1 causes long-range changes in the
conformation of H44 and leads to movement of the domains of
30S with respect to each other. The structure explains how
localized changes at the ribosomal A site lead to global
alterations in the conformation of the 30S subunit.