Summary information and primary citation
- PDB-id
- 1hrz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- NMR
- Summary
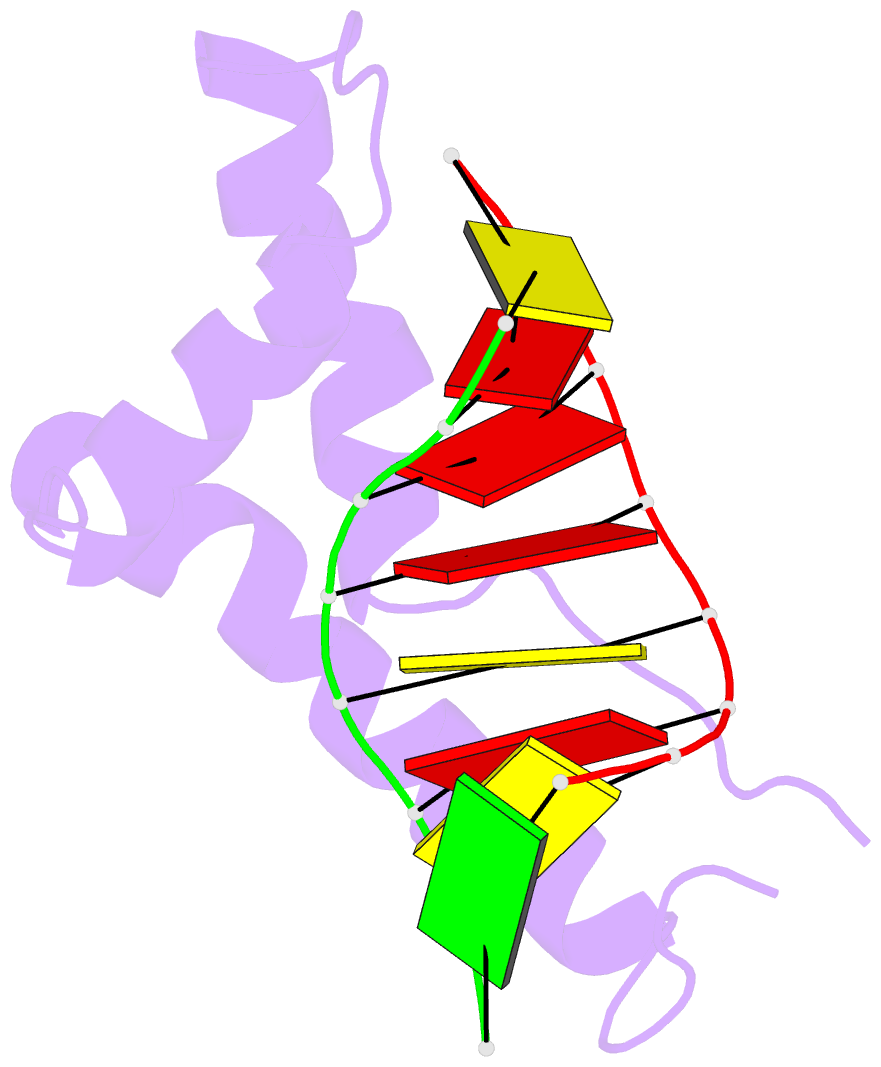
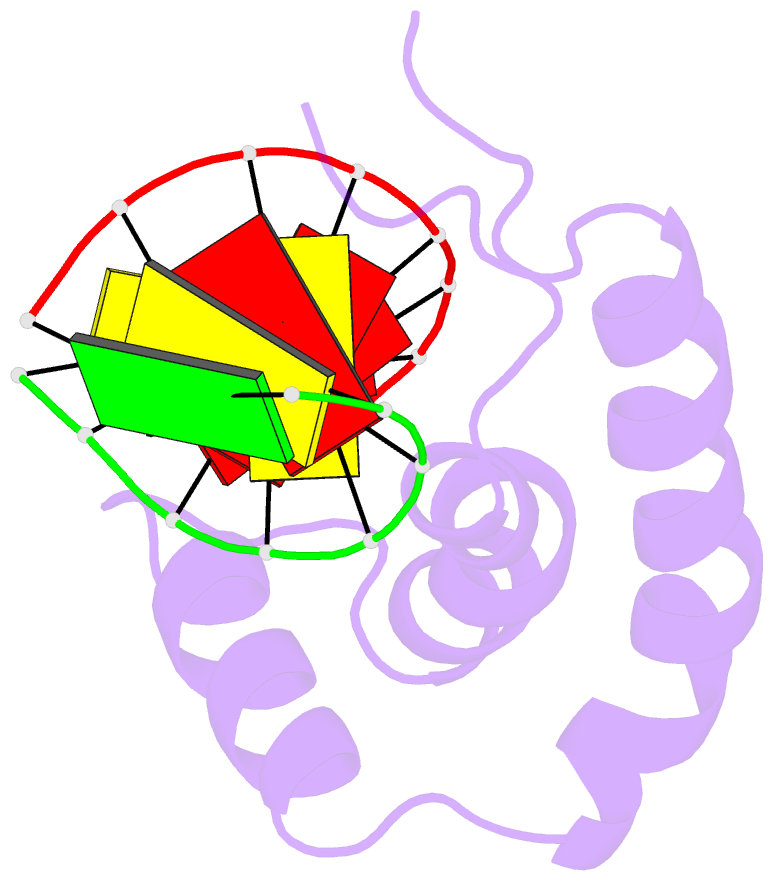
- The 3d structure of the human sry-DNA complex solved by multi-dimensional heteronuclear-edited and-filtered NMR
- Reference
- Werner MH, Huth JR, Gronenborn AM, Clore GM (1995): "Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex." Cell(Cambridge,Mass.), 81, 704-705. doi: 10.1016/0092-8674(95)90532-4.
- Abstract
- The solution structure of the specific complex between the high mobility group (HMG) domain of SRY (hSRY-HMG), the protein encoded by the human testis-determining gene, and its DNA target site in the promoter of the müllerian inhibitory substance gene has been determined by multidimensional NMR spectroscopy. hSRY-HMG has a twisted L shape that presents a concave surface (made up of three helices and the N- and C-terminal strands) to the DNA for sequence-specific recognition. Binding of hSRY-HMG to its specific target site occurs exclusively in the minor groove and induces a large conformational change in the DNA. The DNA in the complex has an overall 70 degrees-80 degrees bend and is helically unwound relative to classical A- and B-DNA. The structure of the complex reveals the origin of sequence-specific binding within the HMG-1/HMG-2 family and provides a framework for understanding the effects of point mutations that cause 46X,Y sex reversal at the atomic level.