Summary information and primary citation
- PDB-id
- 1hut; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-hydrolase inhibitor-DNA
- Method
- X-ray (2.9 Å)
- Summary
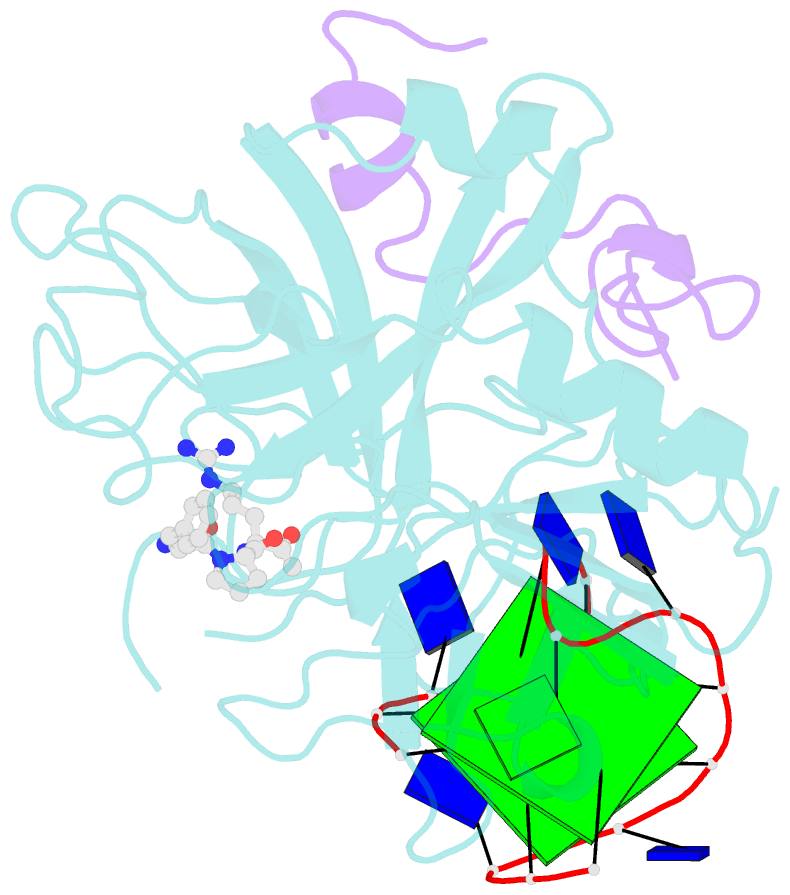
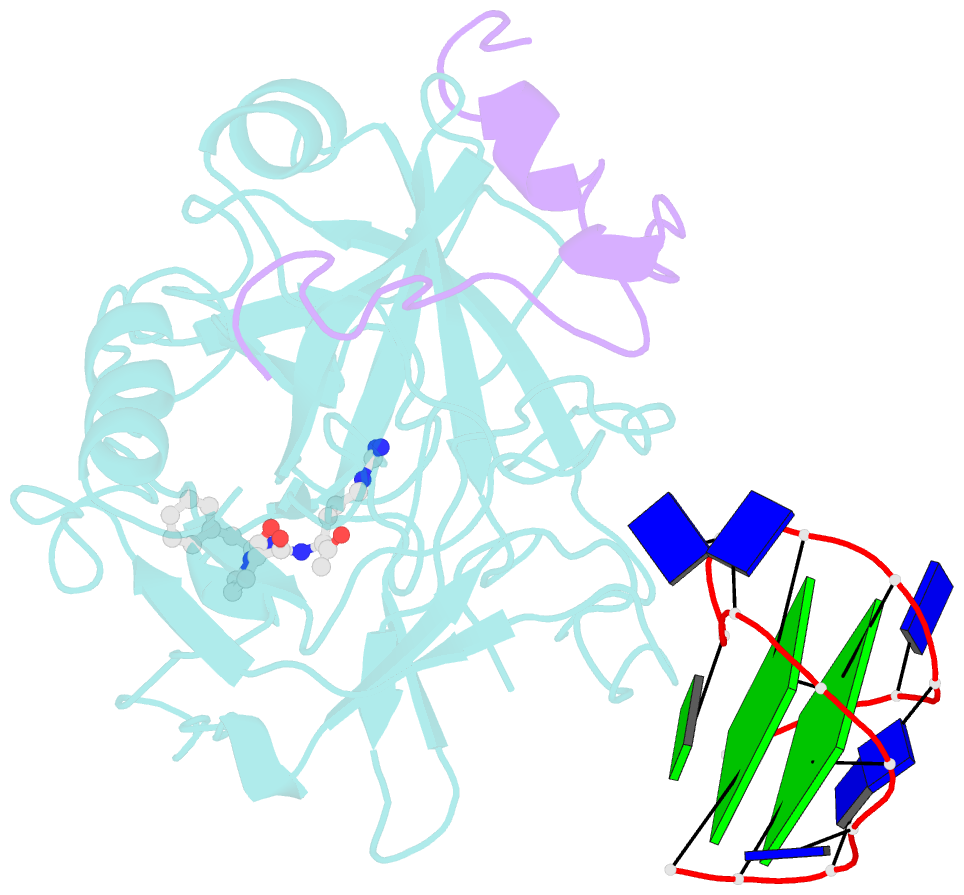
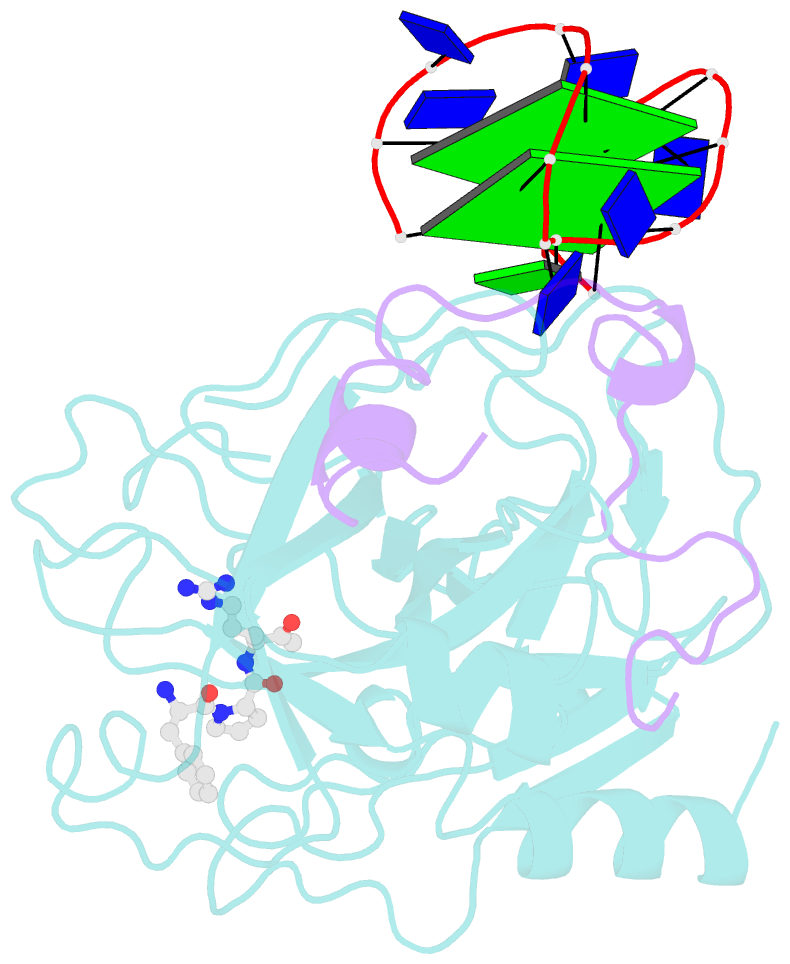
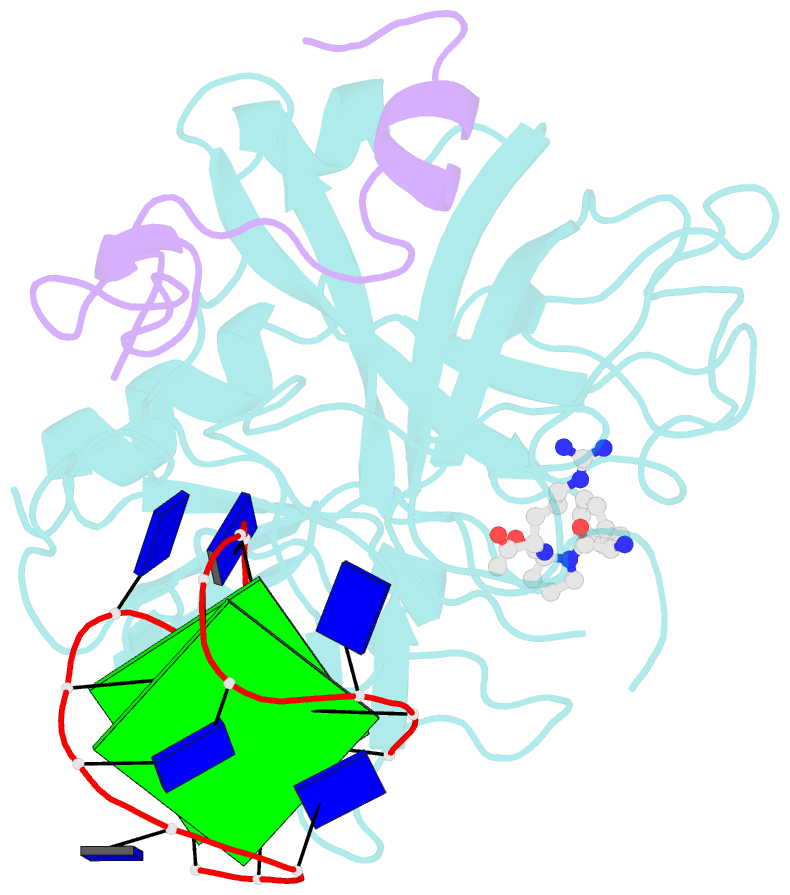
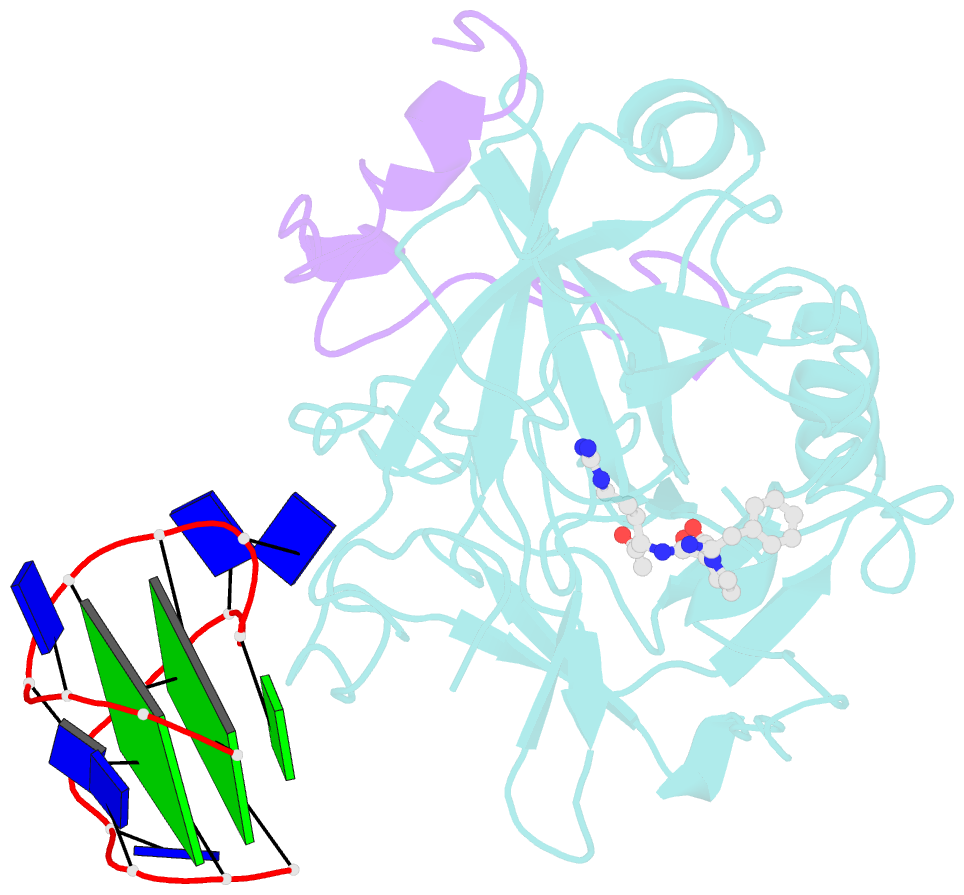
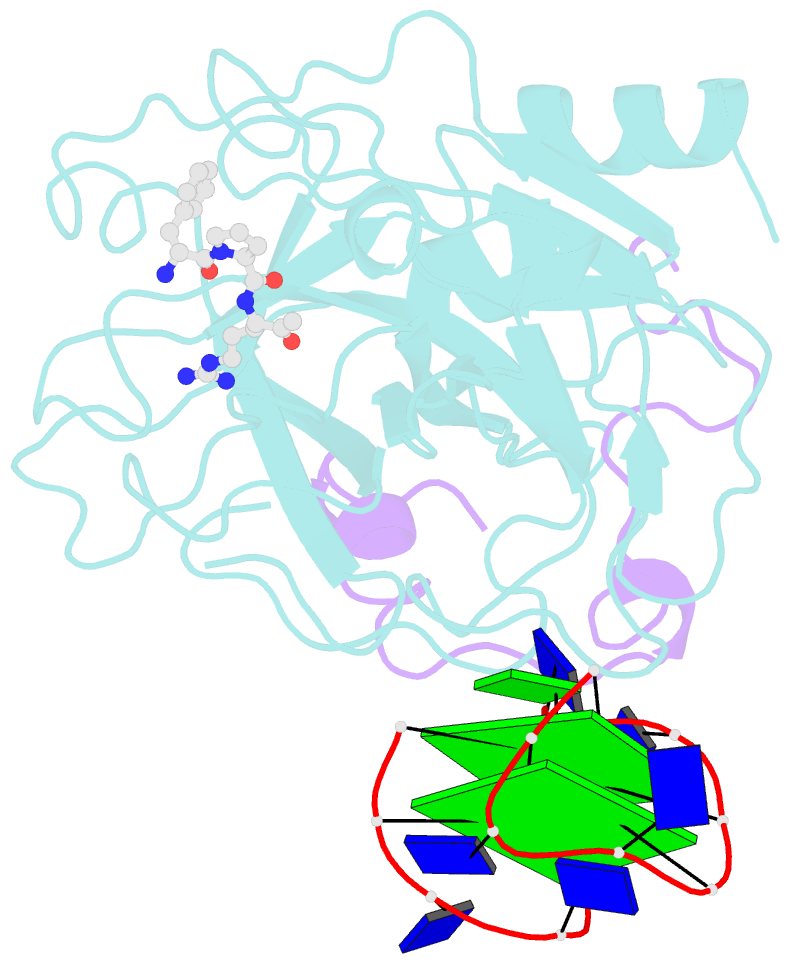
- The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer
- Reference
- Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A (1993): "The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer." J.Biol.Chem., 268, 17651-17654.
- Abstract
- The structure of a complex between human alpha-thrombin and a GGTTGGTGTGGTTGG 15-nucleotide consensus sequence has been solved by x-ray crystallography and refined at 2.9-A resolution to an R value of 0.159. As in solution, in the complex the single-stranded DNA folds into a structure with two G-quartets. The DNA is sandwiched between two different positively charged regions of two symmetry-related thrombin molecules in the crystal structure making ionic and hydrophobic interactions. One region is the fibrinogen recognition exosite and the other, the putative heparin binding site. The lack of inhibition of fibrinogen clotting and platelet activation by the DNA 15-mer with the Arg75-->Glu mutant of thrombin is consistent with the several salt bridges of the DNA in the fibrinogen exosite. The association of DNA with the heparin site of a neighboring molecule appears to simply compensate residual charge. Differences in the 15-mer loop conformations between the complex and NMR solution structures can be attributed to conformational changes upon thrombin binding. Although G-quadruplexes are favored in the presence of monovalent cations, there is no evidence of the latter in the thrombin complex.