Summary information and primary citation
- PDB-id
- 1huz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.6 Å)
- Summary
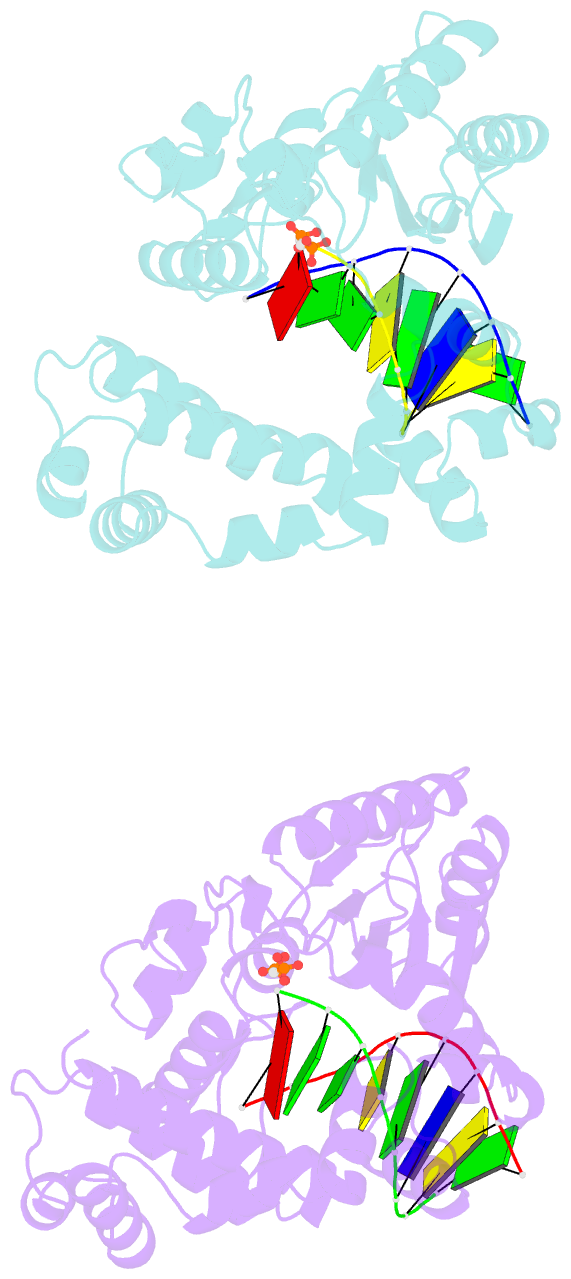
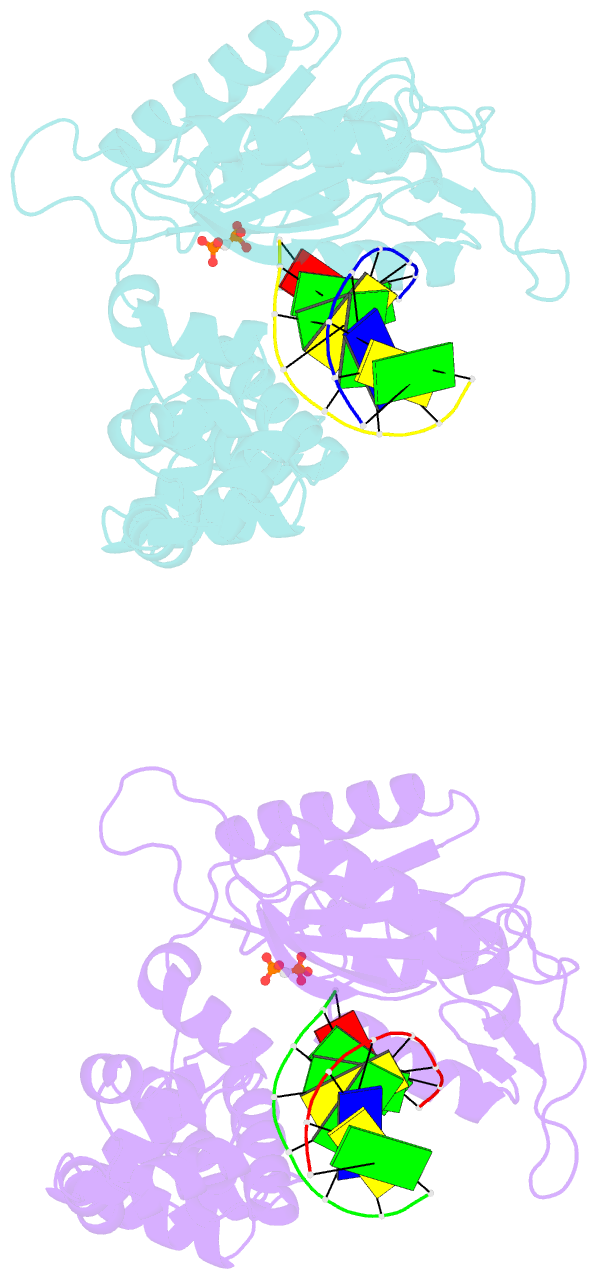
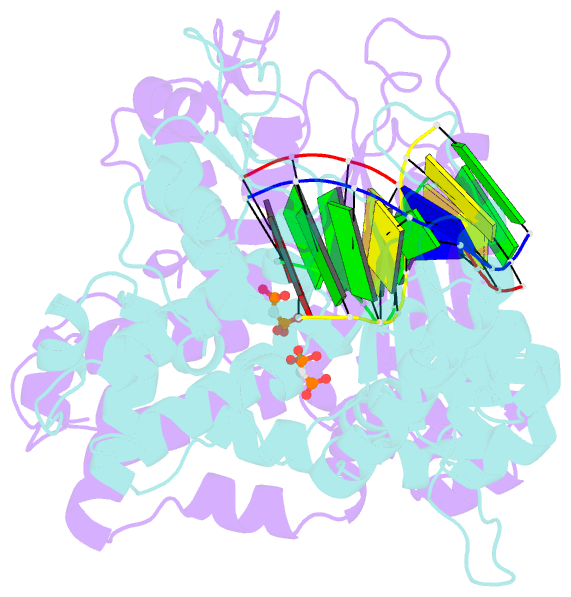
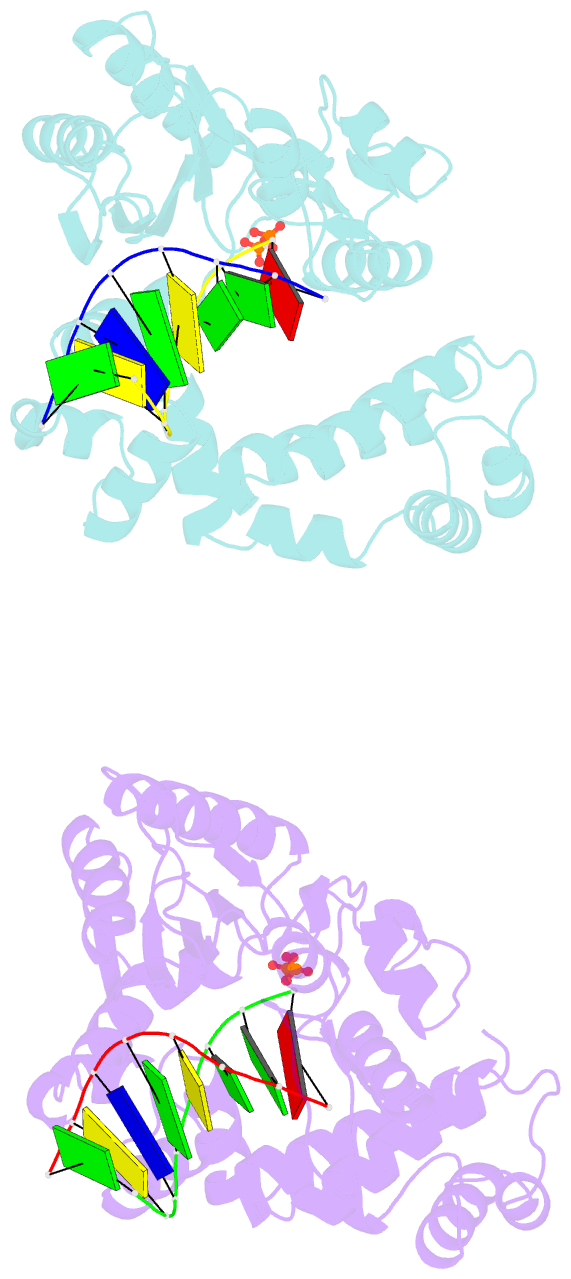
- Crystal structure of DNA polymerase complexed with DNA and cr-pcp
- Reference
- Arndt JW, Gong W, Zhong X, Showalter AK, Liu J, Dunlap CA, Lin Z, Paxson C, Tsai MD, Chan MK (2001): "Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes." Biochemistry, 40, 5368-5375. doi: 10.1021/bi002176j.
- Abstract
- The catalytic reaction mediated by DNA polymerases is known to require two Mg(II) ions, one associated with dNTP binding and the other involved in metal ion catalysis of the chemical step. Here we report a functional intermediate structure of a DNA polymerase with only one metal ion bound, the DNA polymerase beta-DNA template-primer-chromium(III).2'-deoxythymidine 5'-beta,gamma-methylenetriphosphate [Cr(III).dTMPPCP] complex, at 2.6 A resolution. The complex is distinct from the structures of other polymerase-DNA-ddNTP complexes in that the 3'-terminus of the primer has a free hydroxyl group. Hence, this structure represents a fully functional intermediate state. Support for this contention is provided by the observation of turnover in biochemical assays of crystallized protein as well as from the determination that soaking Pol beta crystals with Mn(II) ions leads to formation of the product complex, Pol beta-DNA-Cr(III).PCP, whose structure is also reported. An important feature of both structures is that the fingers subdomain is closed, similar to structures of other ternary complexes in which both metal ion sites are occupied. These results suggest that closing of the fingers subdomain is induced specifically by binding of the metal-dNTP complex prior to binding of the catalytic Mg(2+) ion. This has led us to reevaluate our previous evidence regarding the existence of a rate-limiting conformational change in Pol beta's reaction pathway. The results of stopped-flow studies suggest that there is no detectable rate-limiting conformational change step.