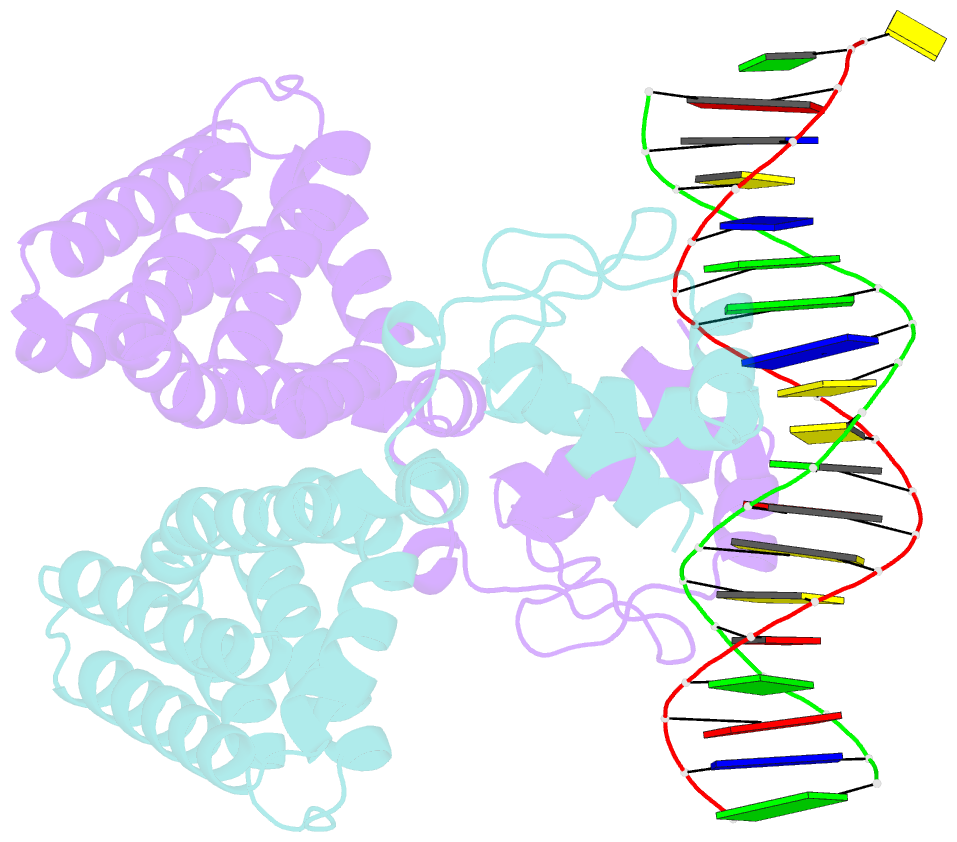
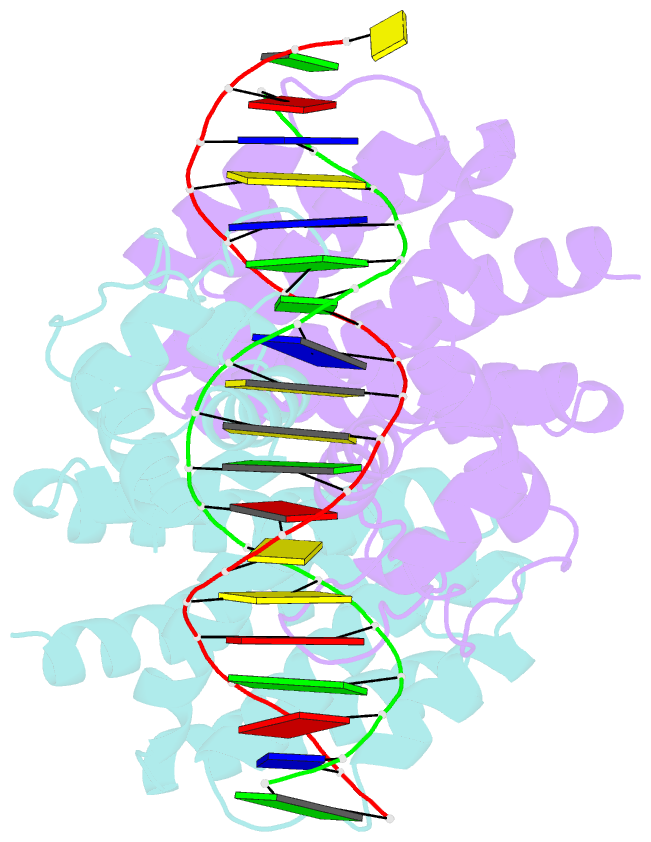
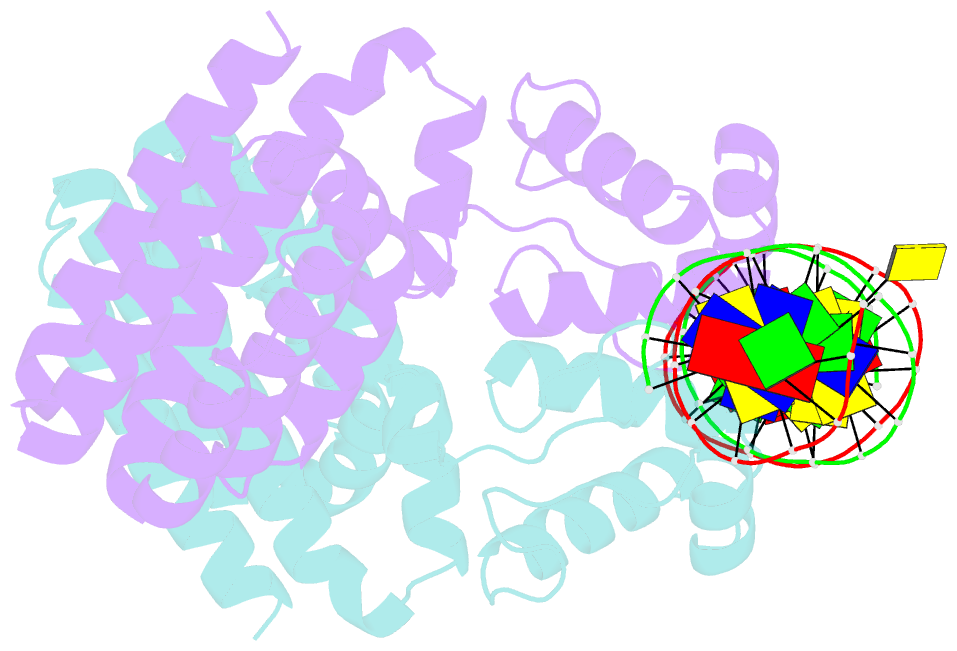
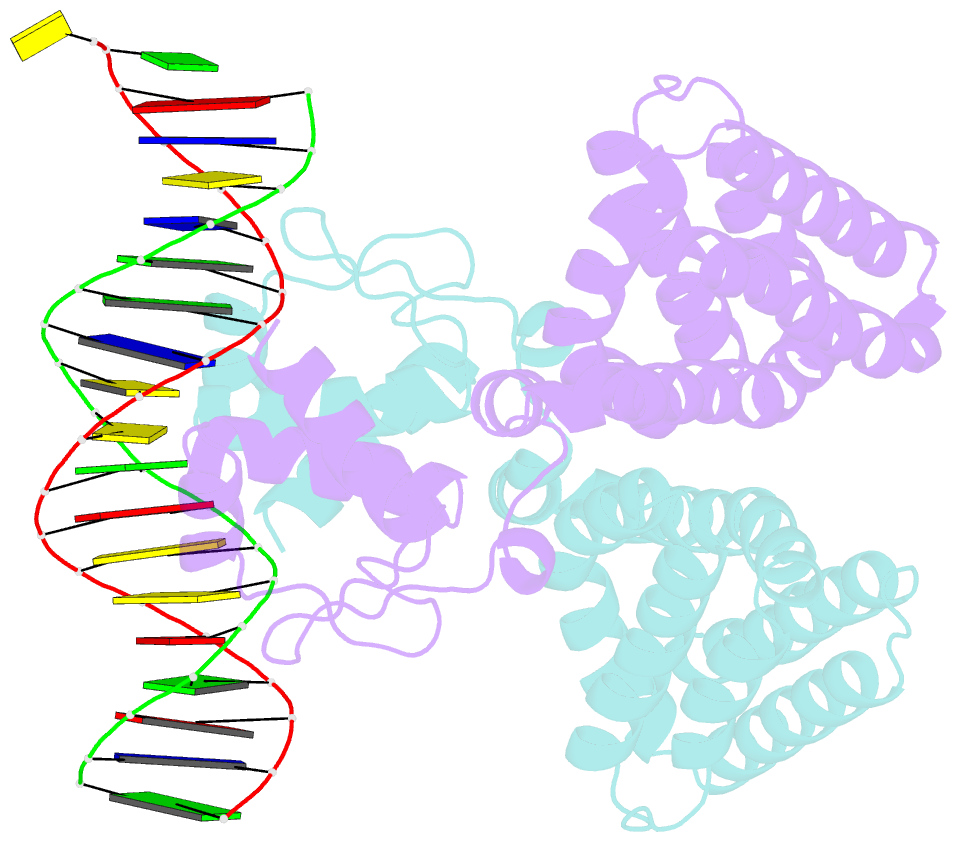
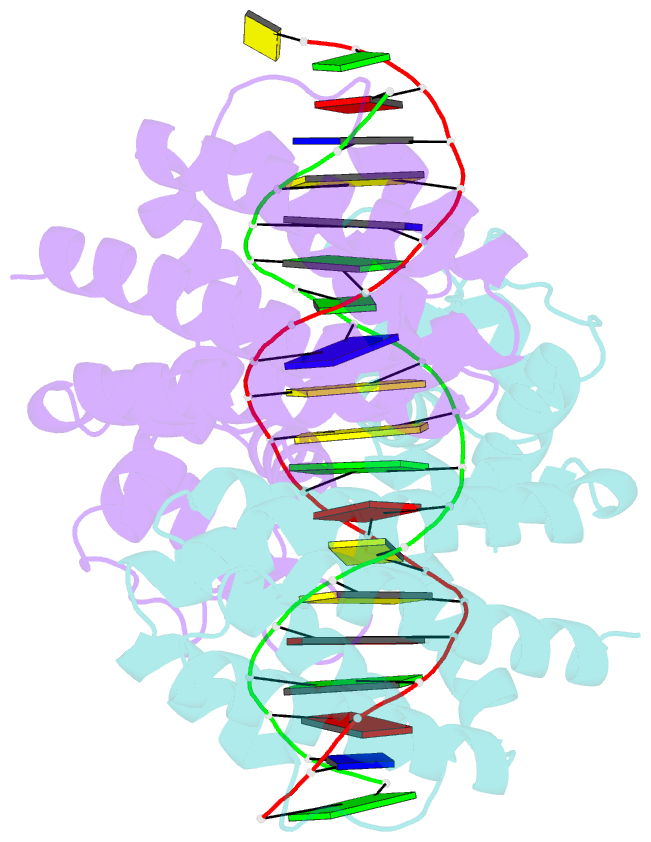
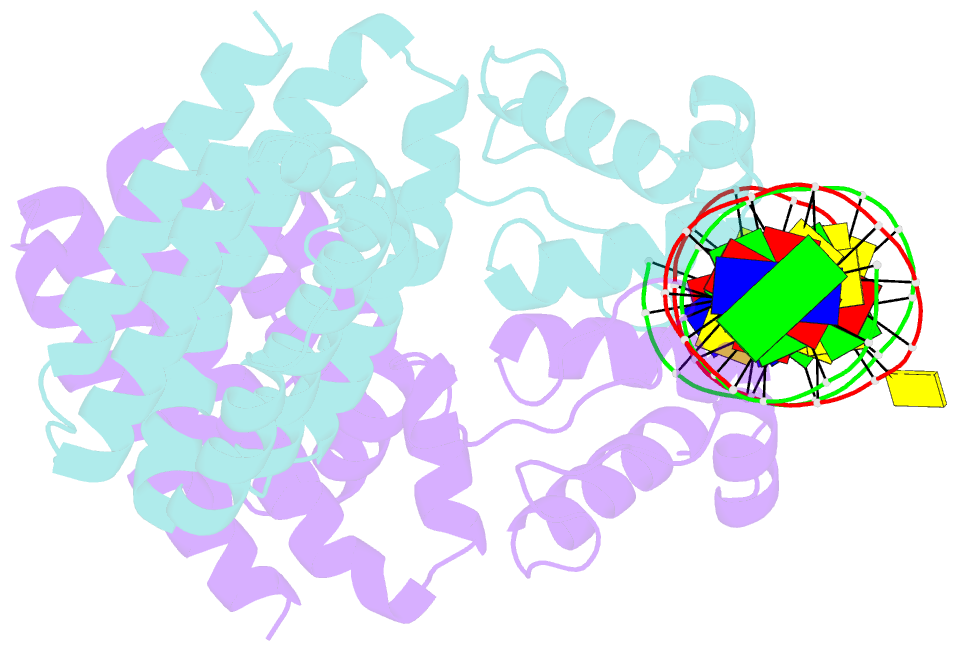
Summary information and primary citation
- PDB-id
- 1hw2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.25 Å)
- Summary
- Fadr-DNA complex: transcriptional control of fatty acid metabolism in echerichia coli
- Reference
- Xu Y, Heath RJ, Li Z, Rock CO, White SW (2001): "The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli." J.Biol.Chem., 276, 17373-17379. doi: 10.1074/jbc.M100195200.
- Abstract
- In Escherichia coli, the expression of fatty acid metabolic genes is controlled by the transcription factor, FadR. The affinity of FadR for DNA is controlled by long chain acyl-CoA molecules, which bind to the protein and modulate gene expression. The crystal structure of FadR reveals a two domain dimeric molecule where the N-terminal domains bind DNA, and the C-terminal domains bind acyl-CoA. The DNA binding domain has a winged-helix motif, and the C-terminal domain resembles the sensor domain of the Tet repressor. The FadR.DNA complex reveals how the protein interacts with DNA and specifically recognizes a palindromic sequence. Structural and functional similarities to the Tet repressor and the BmrR transcription factors suggest how the binding of the acyl-CoA effector molecule to the C-terminal domain may affect the DNA binding affinity of the N-terminal domain. We suggest that the binding of acyl-CoA disrupts a buried network of charged and polar residues in the C-terminal domain, and the resulting conformational change is transmitted to the N-terminal domain via a domain-spanning alpha-helix.