Summary information and primary citation
- PDB-id
- 1hwt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation-DNA
- Method
- X-ray (2.5 Å)
- Summary
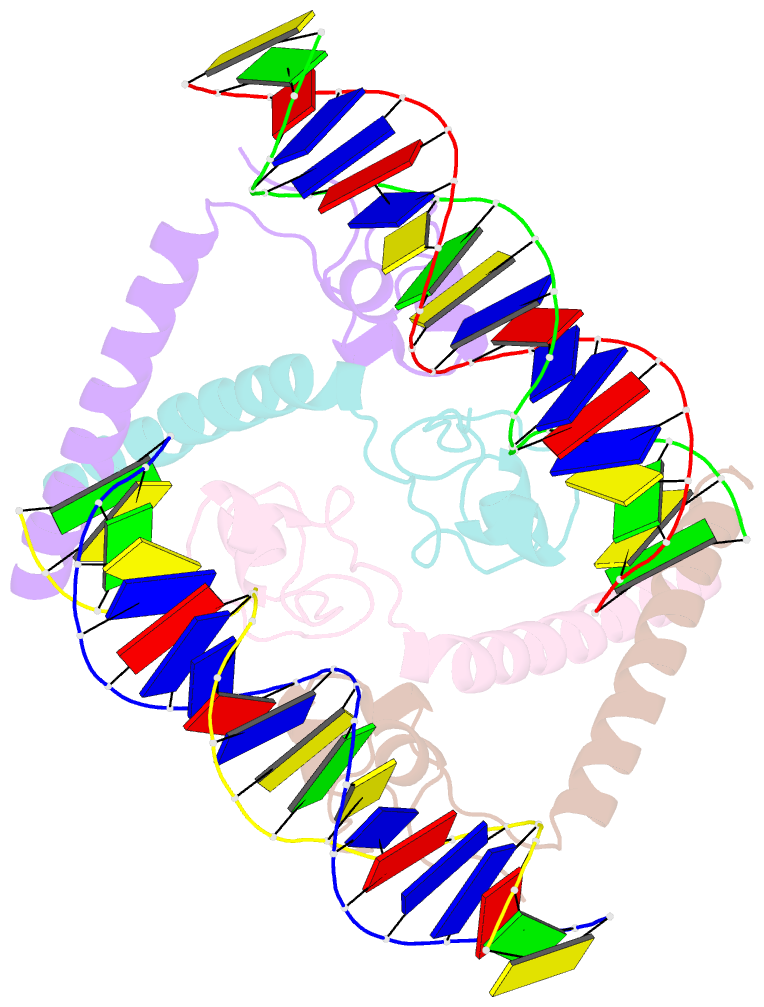
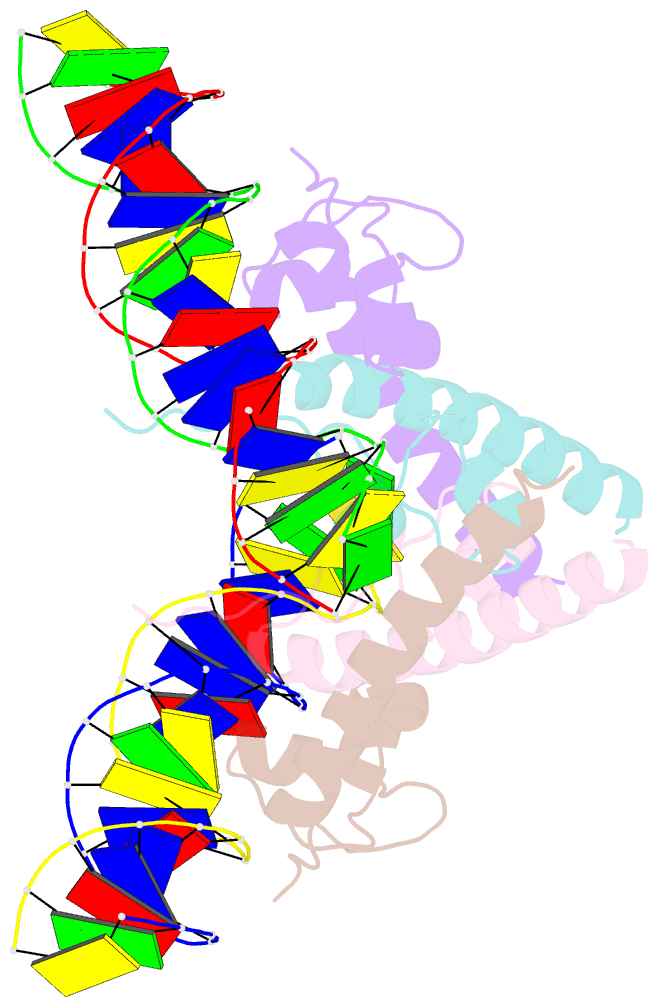
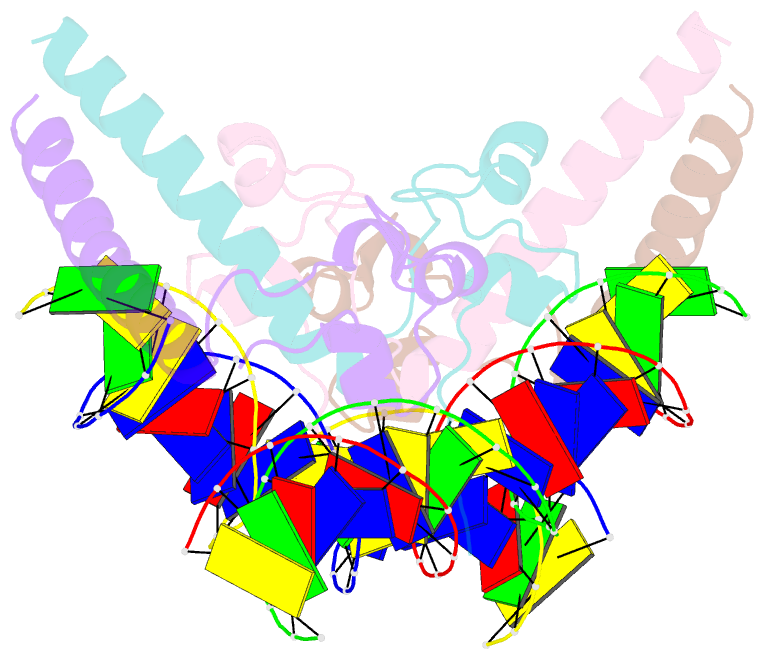
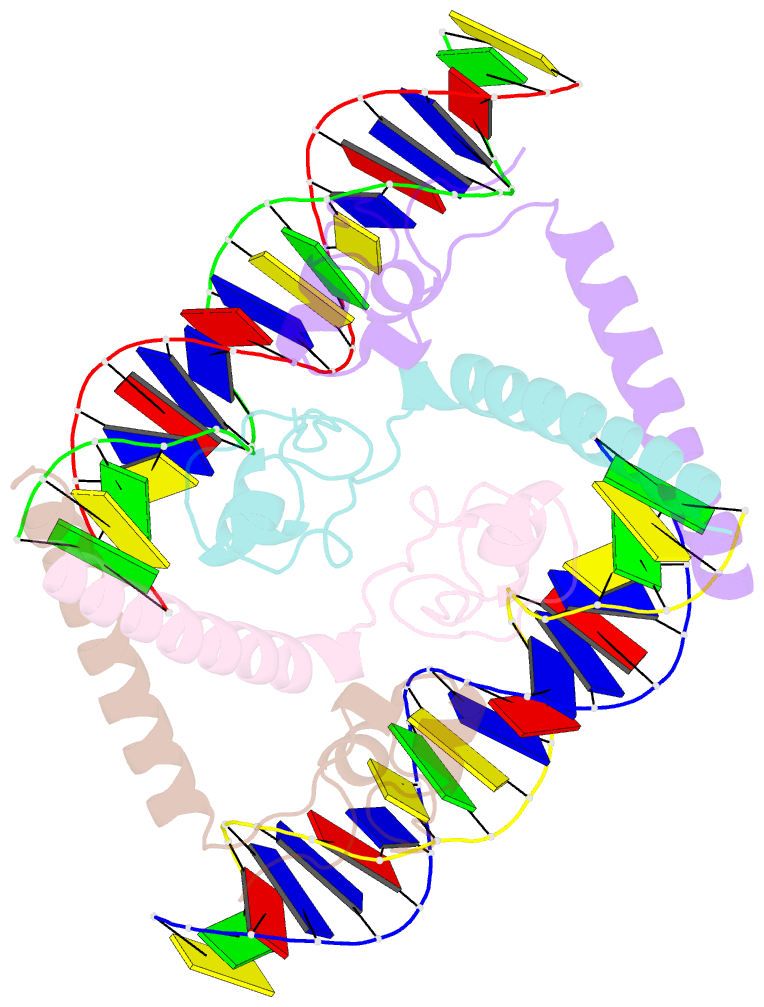
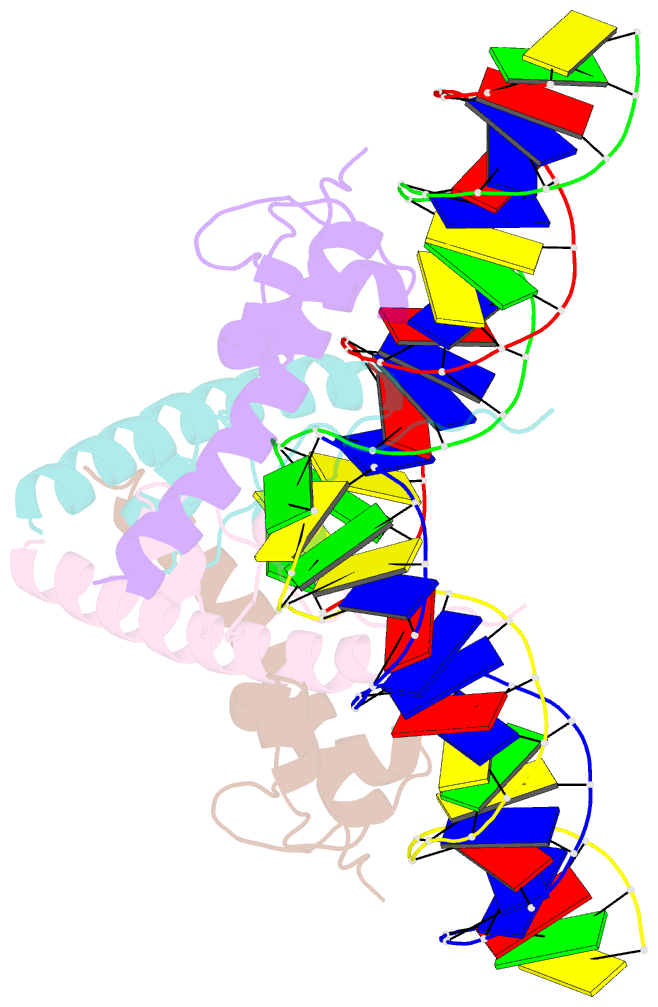
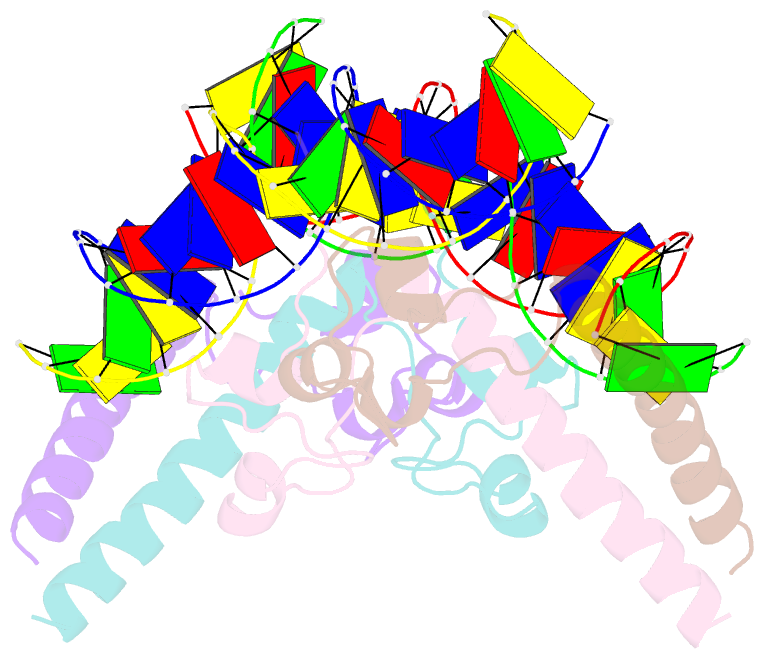
- Structure of a hap1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein
- Reference
- King DA, Zhang L, Guarente L, Marmorstein R (1999): "Structure of a HAP1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein." Nat.Struct.Biol., 6, 64-71. doi: 10.1038/4940.
- Abstract
- HAP1 is a member of a family of fungal transcription factors that contain a Zn2Cys6 binuclear cluster domain and bind as homodimers to sequences containing two DNA half sites. We have determined the 2.5 A crystal structure of HAP1 bound to a cognate upstream activation sequence from the CYC7 gene. The structure reveals that HAP1 is bound in a dramatically asymmetric manner to the DNA target. This asymmetry aligns the Zn2Cys6 domains in a tandem head-to-tail fashion to contact two DNA half sites, positions an N-terminal arm of one of the protein subunits to interact with the inter-half site base pairs in the DNA minor groove, and suggests a mechanism by which DNA-binding facilitates asymmetric dimerization by HAP1. Comparisons with the DNA complexes of the related GAL4, PPR1 and PUT3 proteins illustrate how a conserved protein domain can be reoriented to recognize DNA half sites of different polarities and how homodimeric proteins adopt dramatically asymmetric structures to recognize cognate DNA targets.