Summary information and primary citation
- PDB-id
- 1i5l; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.75 Å)
- Summary
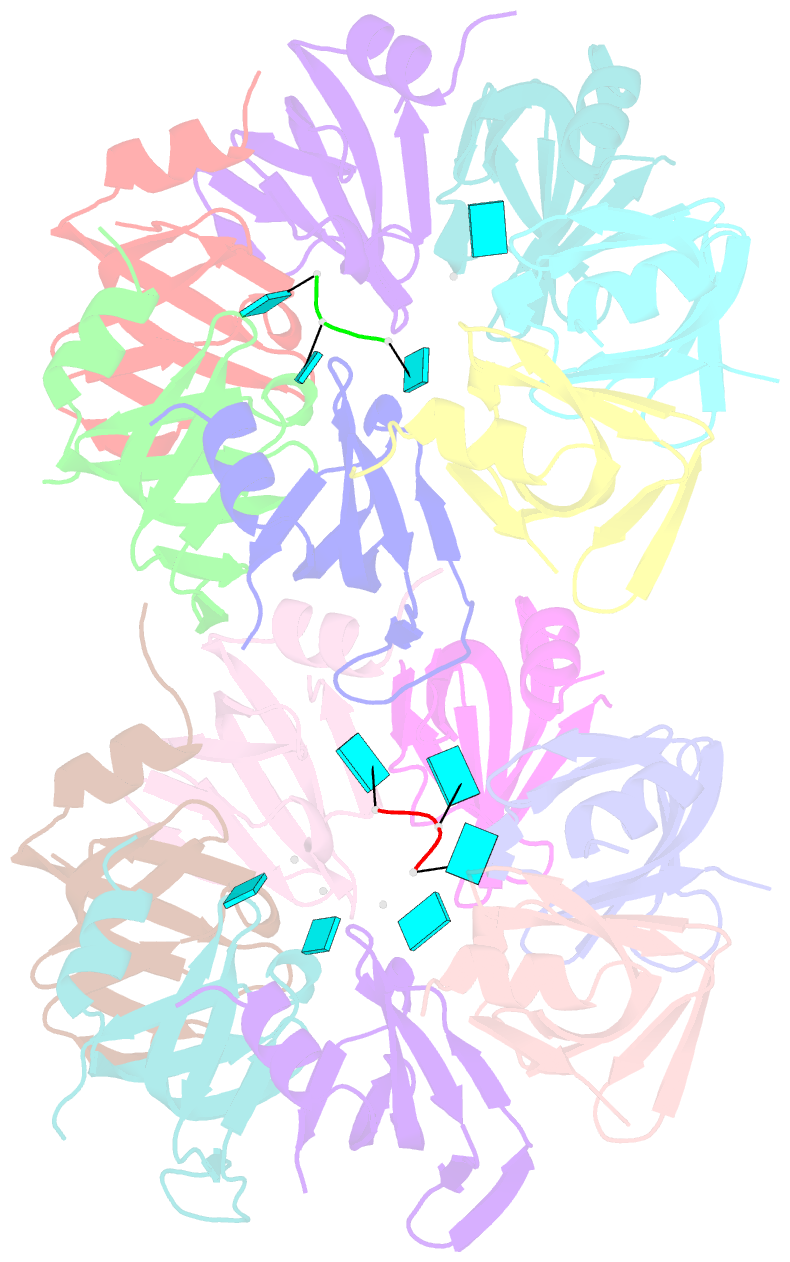
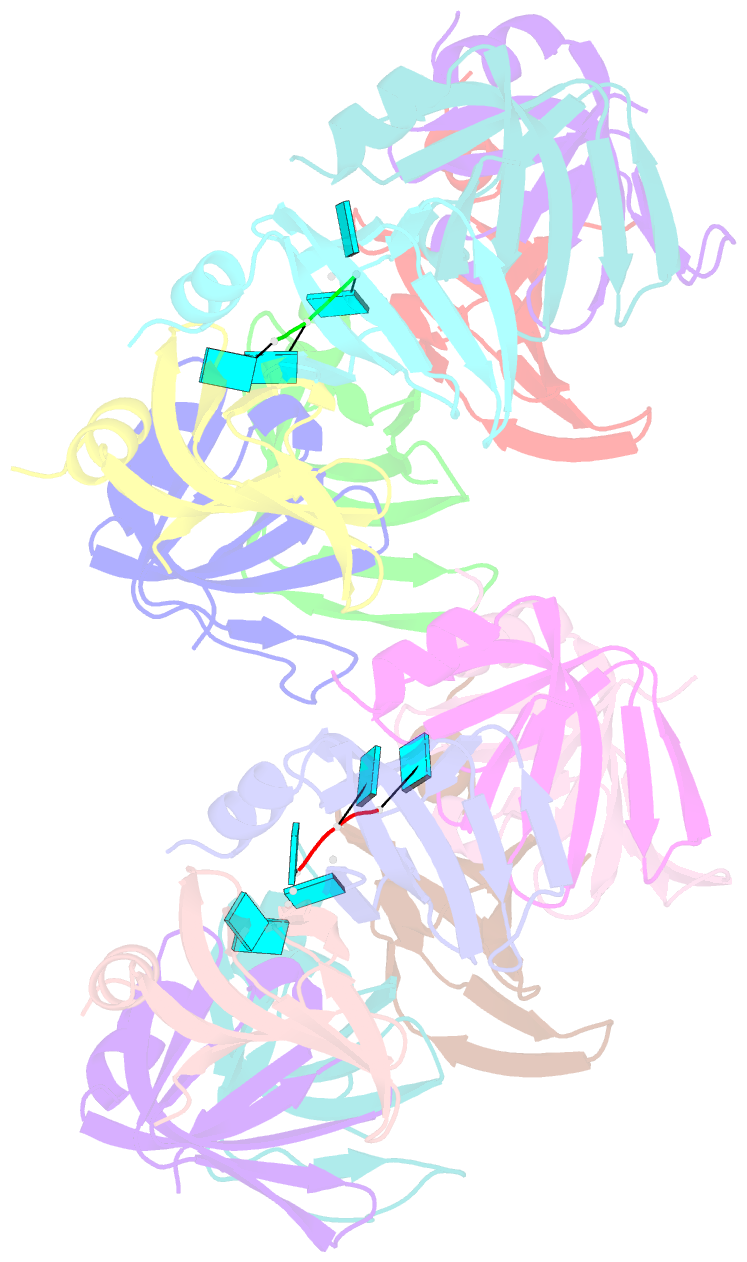
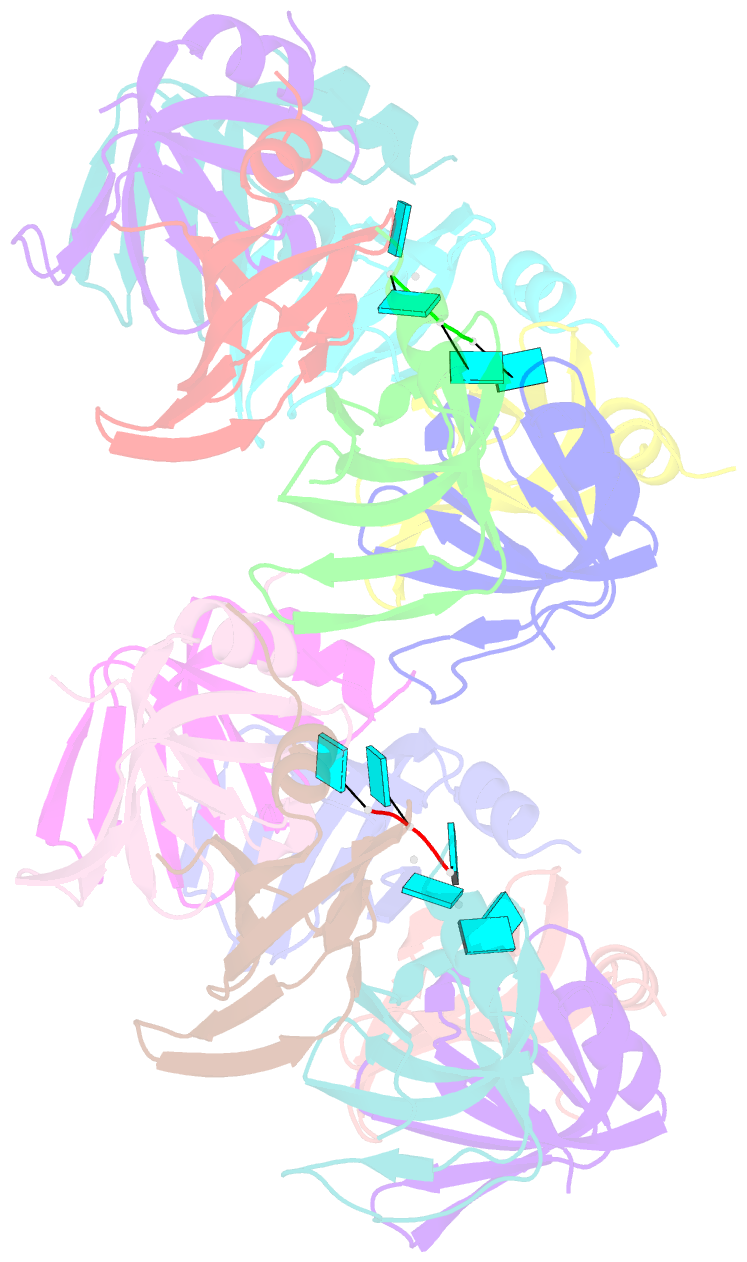
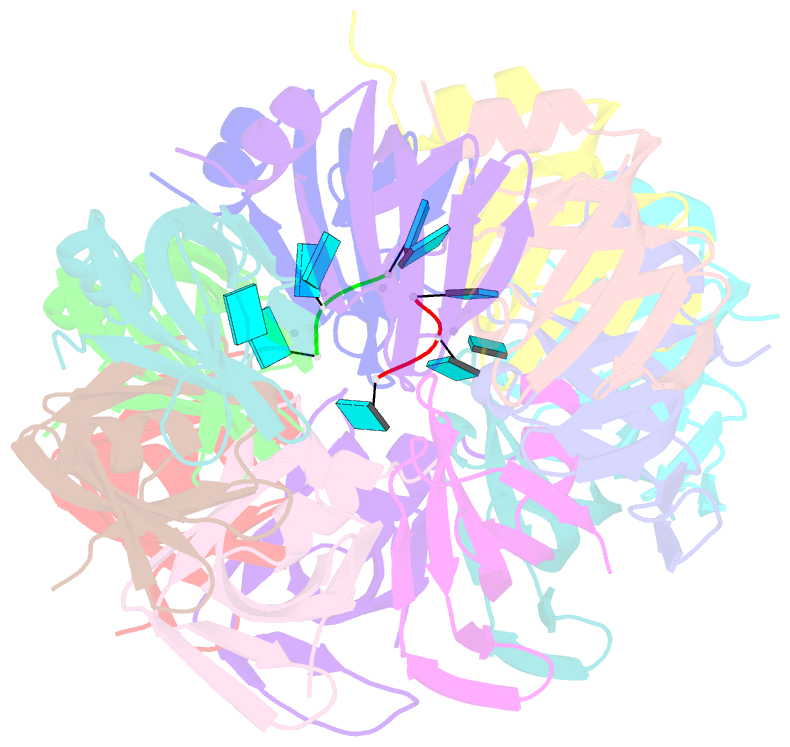
- Crystal structure of an sm-like protein (af-sm1) from archaeoglobus fulgidus complexed with short poly-u RNA
- Reference
- Toro I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D (2001): "RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex." EMBO J., 20, 2293-2303. doi: 10.1093/emboj/20.9.2293.
- Abstract
- Eukaryotic Sm and Sm-like proteins associate with RNA to form the core domain of ribonucleoprotein particles involved in pre-mRNA splicing and other processes. Recently, putative Sm proteins of unknown function have been identified in Archaea. We show by immunoprecipitation experiments that the two Sm proteins present in Archaeoglobus fulgidus (AF-Sm1 and AF-Sm2) associate with RNase P RNA in vivo, suggesting a role in tRNA processing. The AF-Sm1 protein also interacts specifically with oligouridylate in vitro. We have solved the crystal structures of this protein and a complex with RNA. AF-Sm1 forms a seven-membered ring, with the RNA interacting inside the central cavity on one face of the doughnut-shaped complex. The bases are bound via stacking and specific hydrogen bonding contacts in pockets lined by residues highly conserved in archaeal and eukaryotic Sm proteins, while the phosphates remain solvent accessible. A comparison with the structures of human Sm protein dimers reveals closely related monomer folds and intersubunit contacts, indicating that the architecture of the Sm core domain and RNA binding have been conserved during evolution.