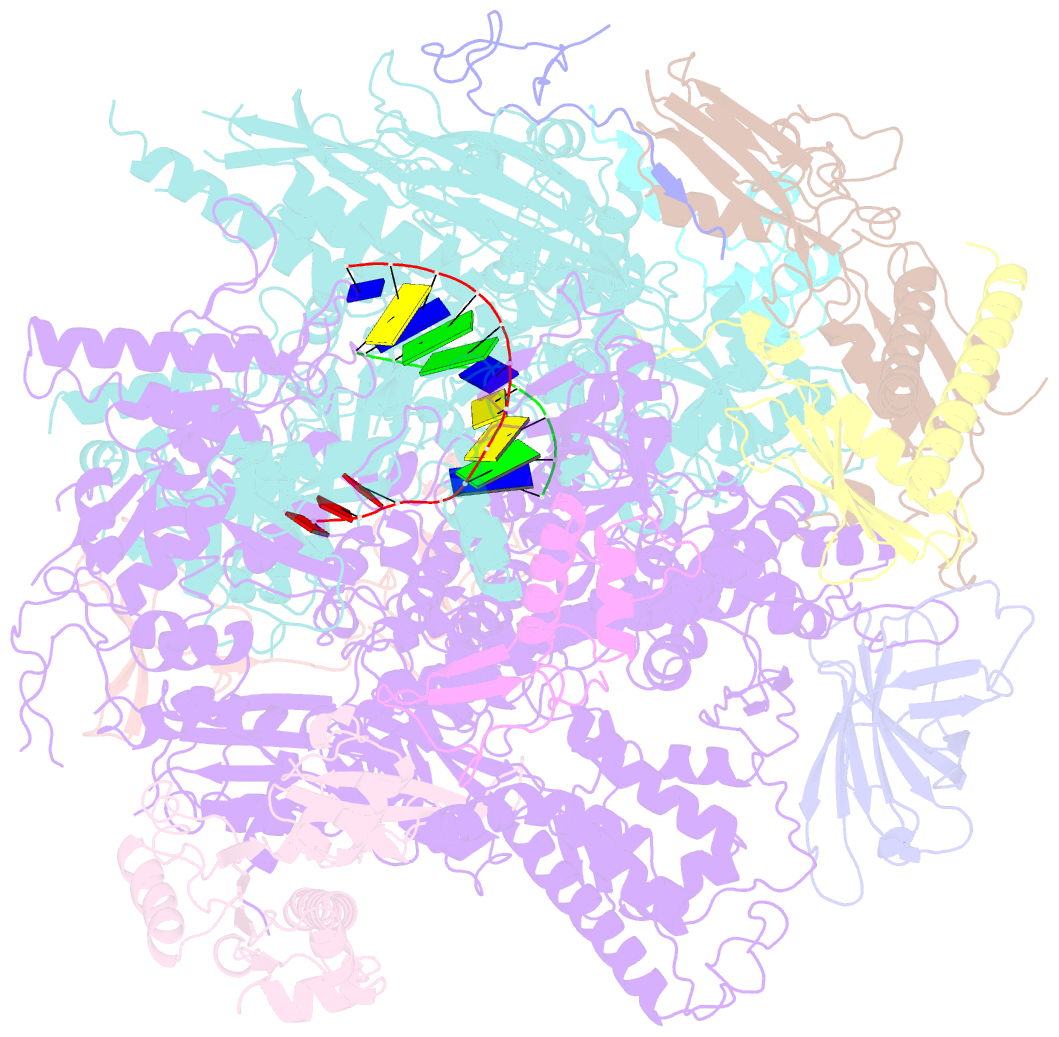
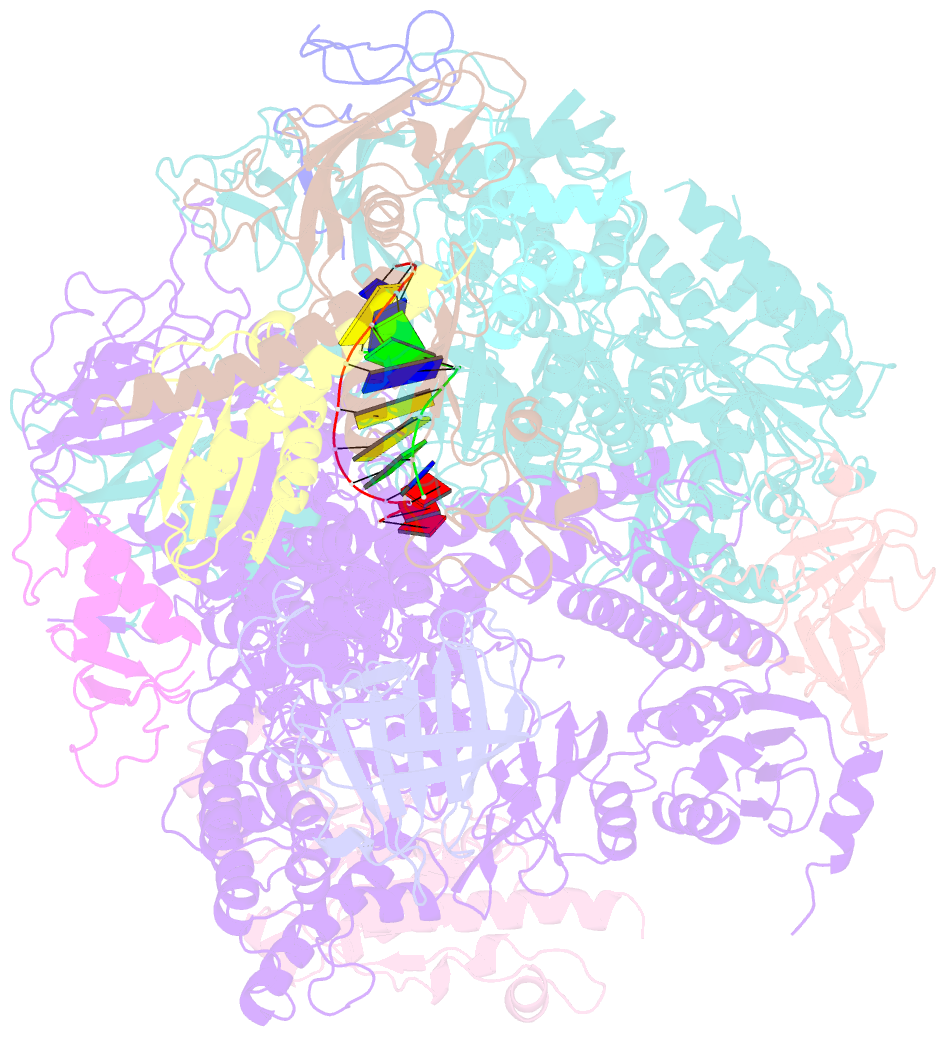
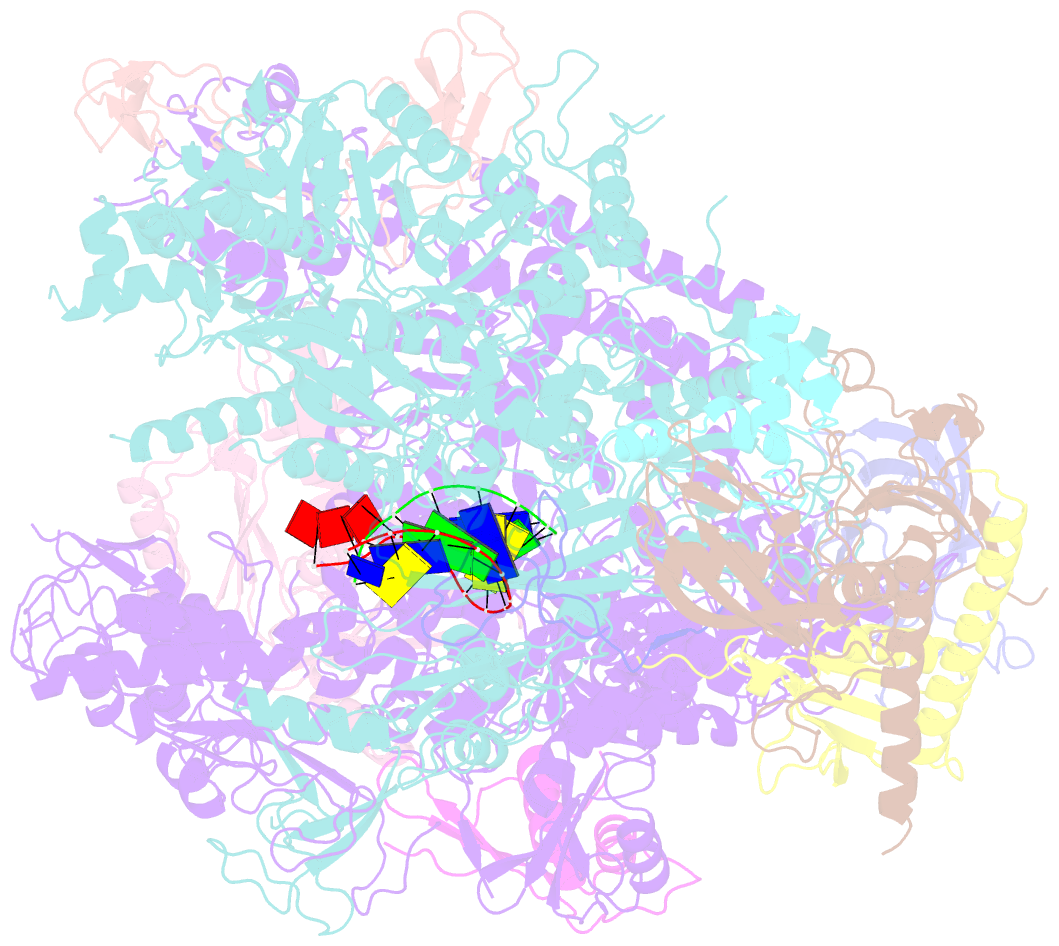
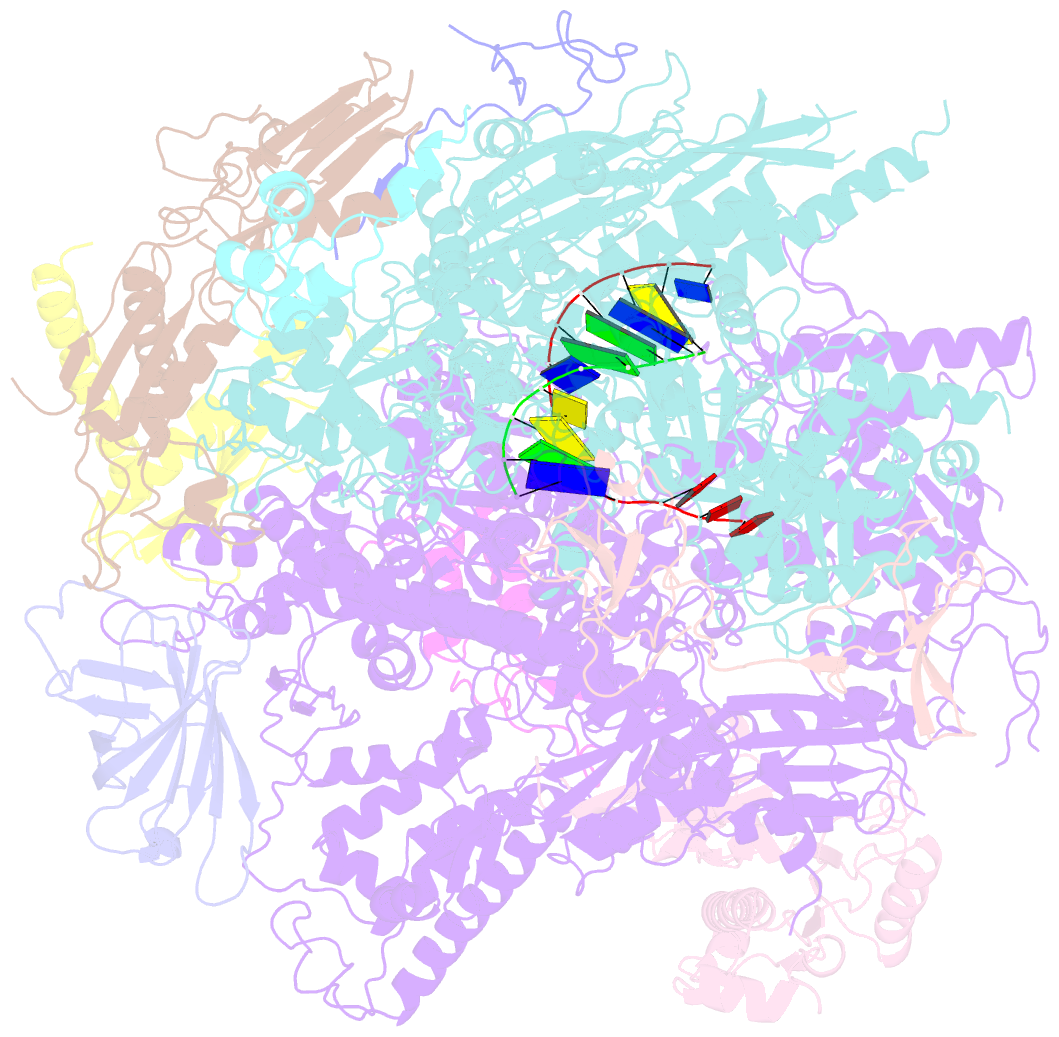
Summary information and primary citation
- PDB-id
- 1i6h; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA-RNA hybrid
- Method
- X-ray (3.3 Å)
- Summary
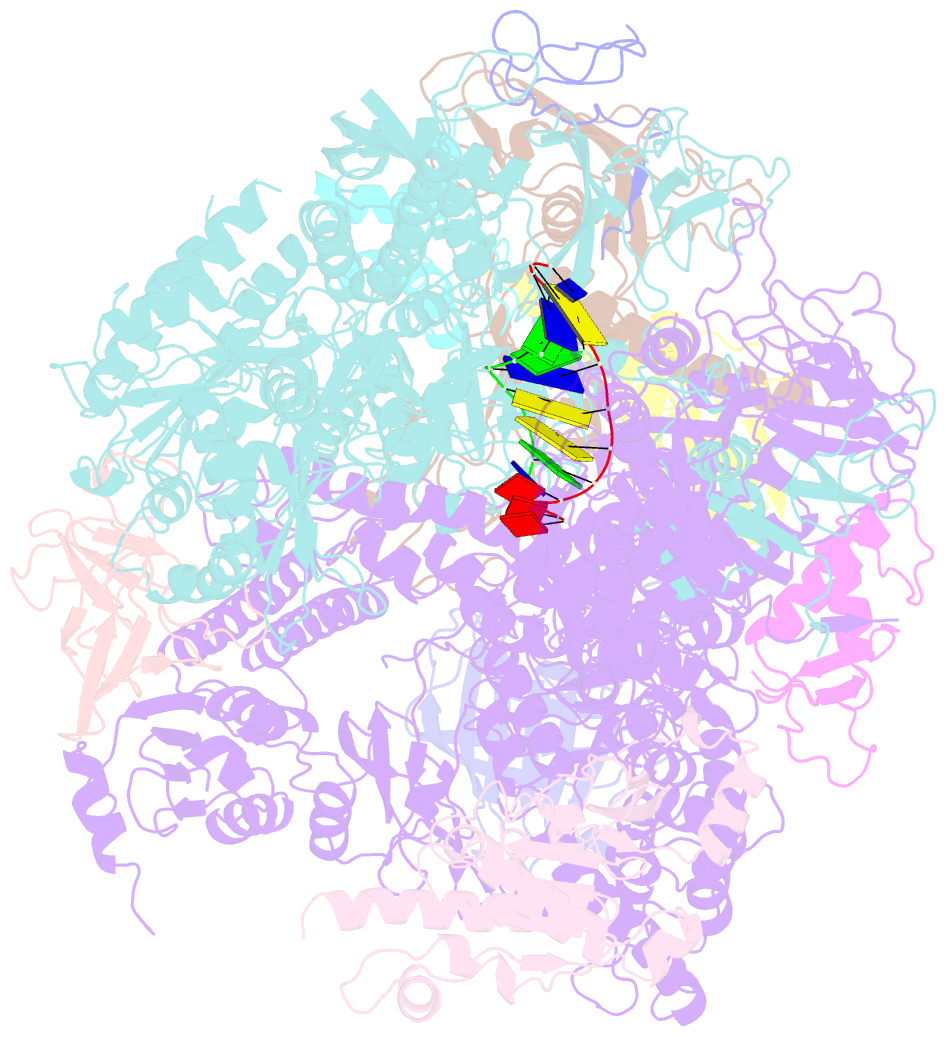
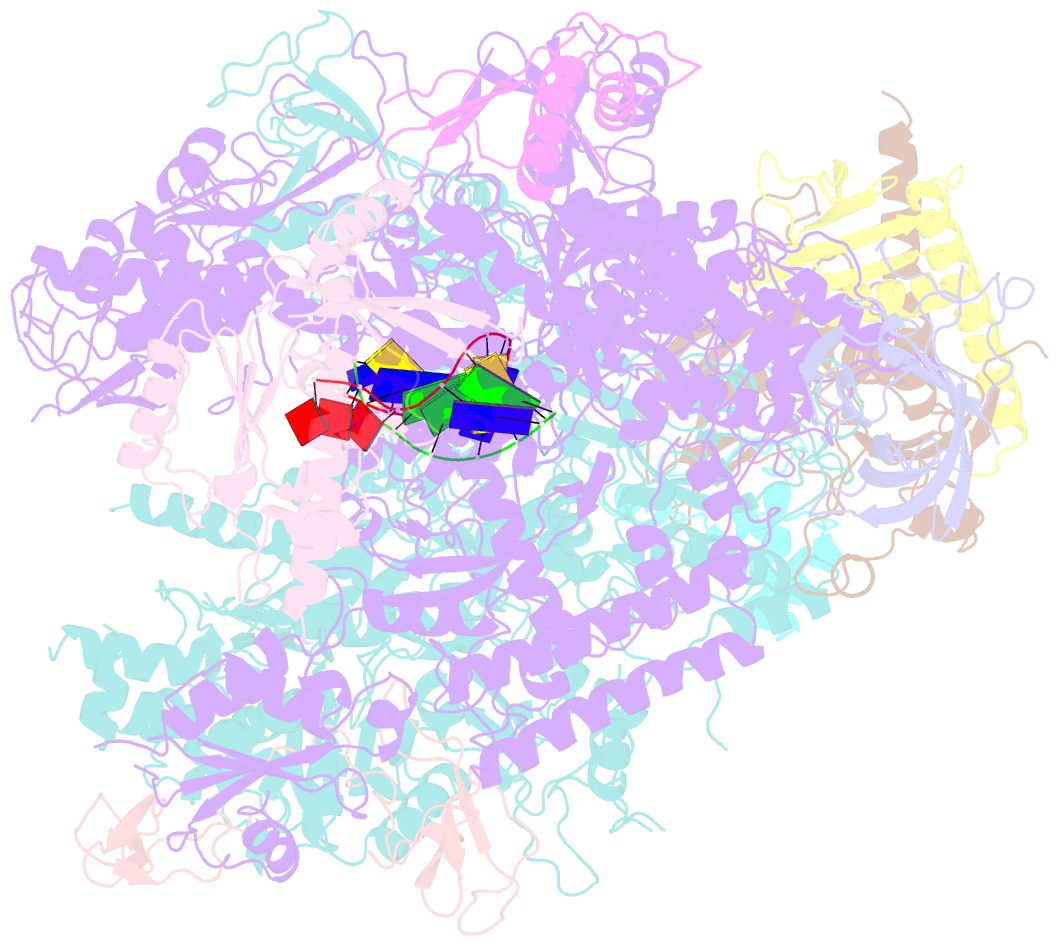
- RNA polymerase ii elongation complex
- Reference
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD (2001): "Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution." Science, 292, 1876-1882. doi: 10.1126/science.1059495.
- Abstract
- The crystal structure of RNA polymerase II in the act of transcription was determined at 3.3 A resolution. Duplex DNA is seen entering the main cleft of the enzyme and unwinding before the active site. Nine base pairs of DNA-RNA hybrid extend from the active center at nearly right angles to the entering DNA, with the 3' end of the RNA in the nucleotide addition site. The 3' end is positioned above a pore, through which nucleotides may enter and through which RNA may be extruded during back-tracking. The 5'-most residue of the RNA is close to the point of entry to an exit groove. Changes in protein structure between the transcribing complex and free enzyme include closure of a clamp over the DNA and RNA and ordering of a series of "switches" at the base of the clamp to create a binding site complementary to the DNA-RNA hybrid. Protein-nucleic acid contacts help explain DNA and RNA strand separation, the specificity of RNA synthesis, "abortive cycling" during transcription initiation, and RNA and DNA translocation during transcription elongation.