Summary information and primary citation
- PDB-id
- 1i6j; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.0 Å)
- Summary
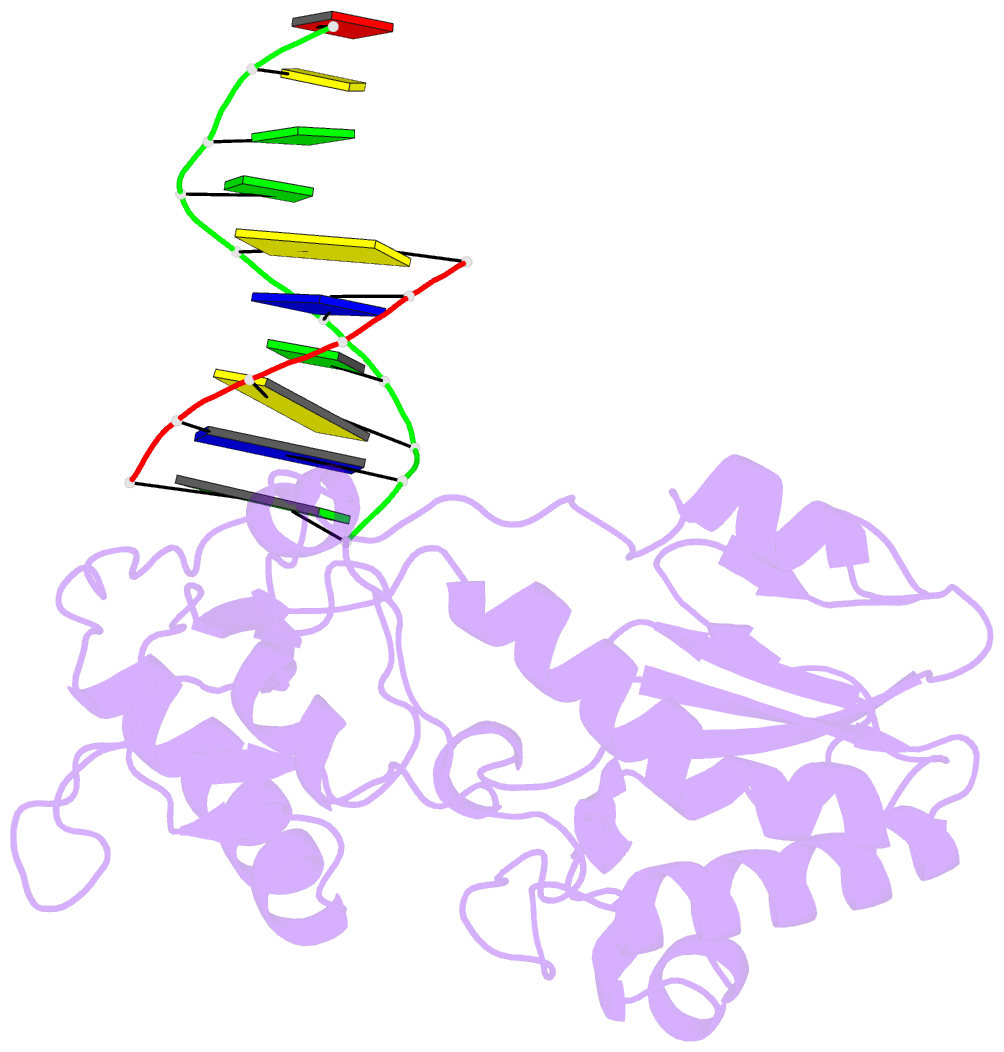
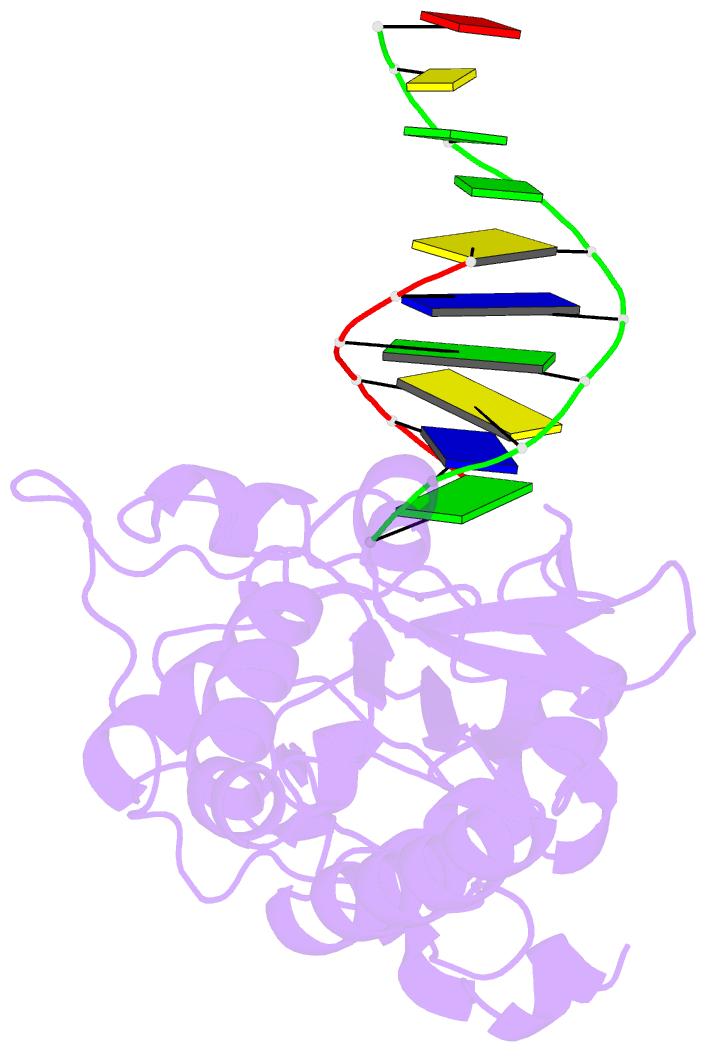
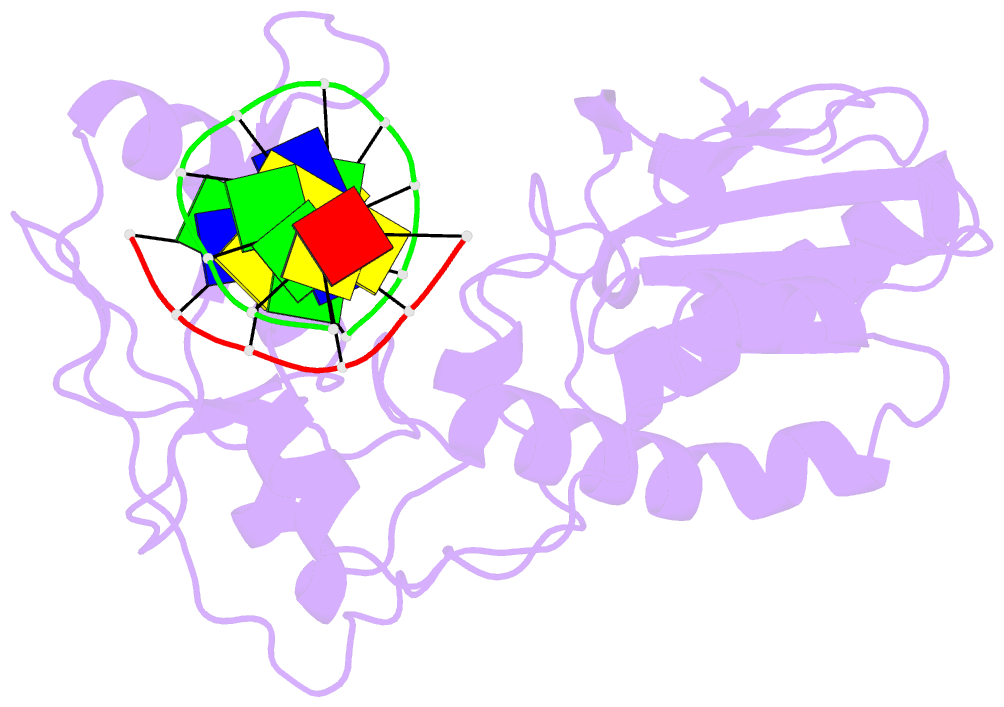
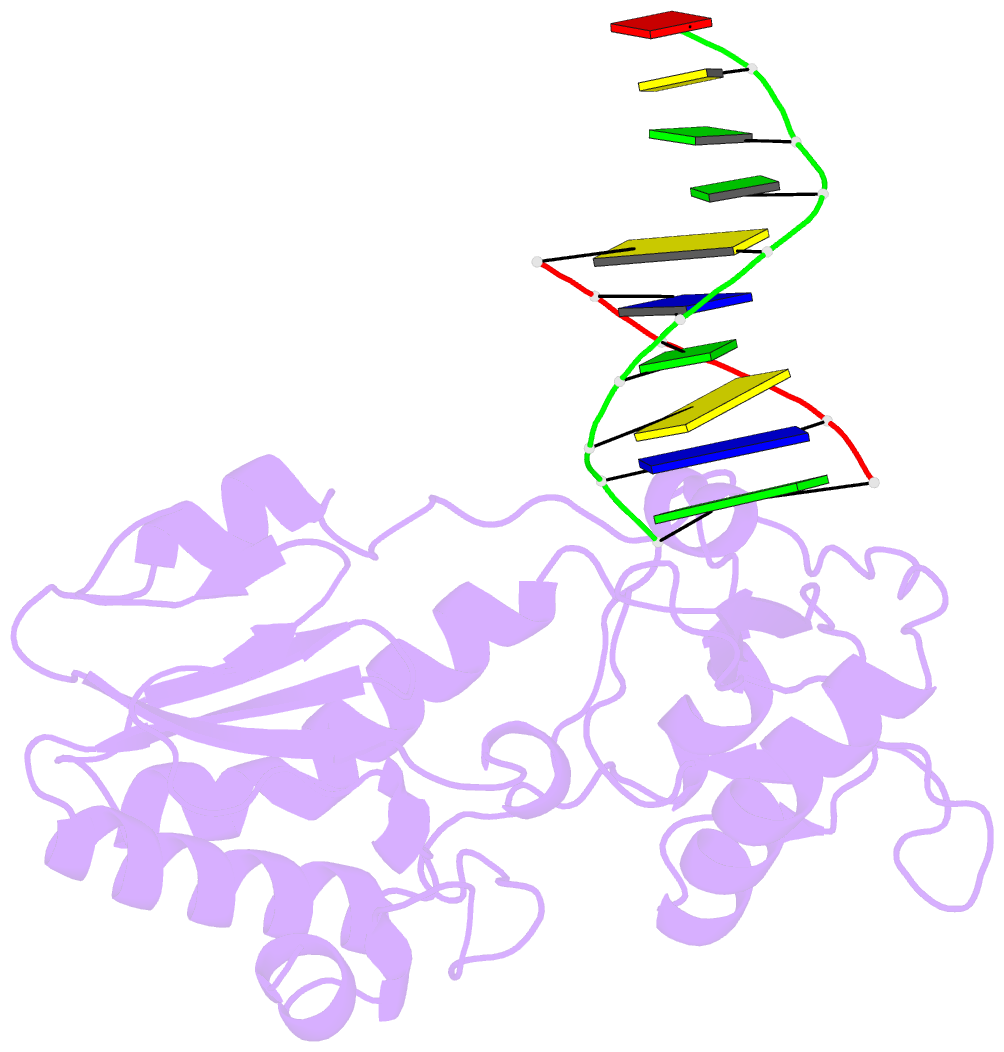
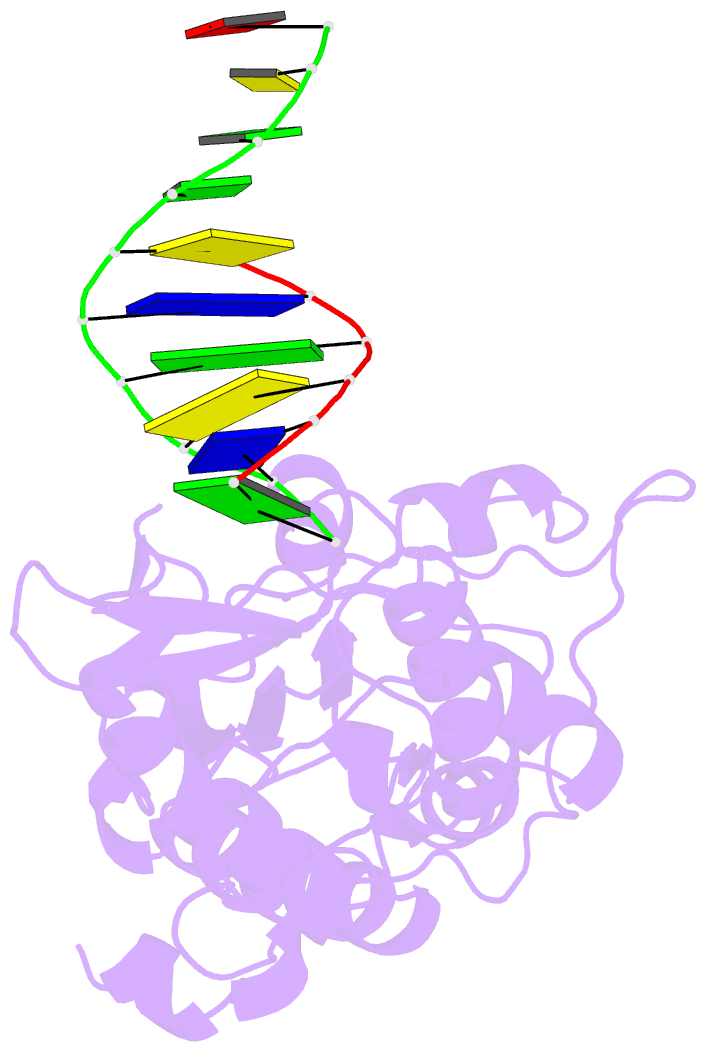
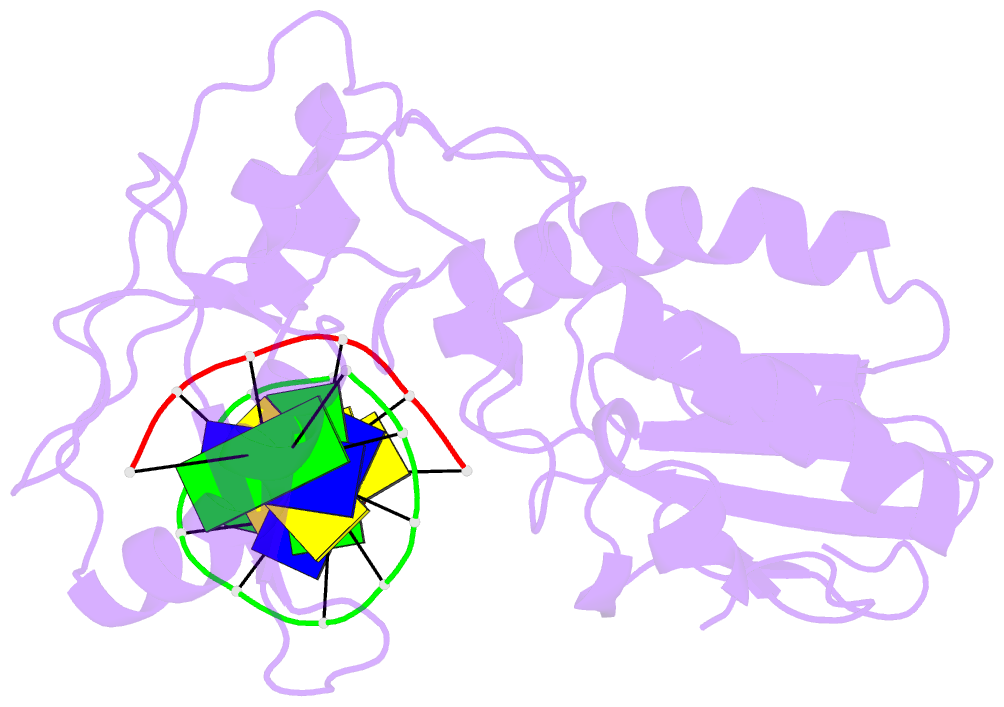
- Crystal structure of a pseudo-16-mer DNA with stacked guanines and two g-a mispairs complexed with the n-terminal fragment of moloney murine leukemia virus reverse transcriptase
- Reference
- Cote ML, Georgiadis MM (2001): "Structure of a pseudo-16-mer DNA with stacked guanines and two G-A mispairs complexed with the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase." Acta Crystallogr.,Sect.D, 57, 1238-1250. doi: 10.1107/S090744490100943X.
- Abstract
- The X-ray crystal structure at 2.0 A resolution of a DNA molecule complexed with the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase (MMLV RT) has been determined. This method allows the study of nucleic acids in a unique and largely unfettered environment without the complicated lattice interactions typically observed in DNA-only crystal structures. Molecular-replacement phasing using only the protein provided readily interpretable electron density with no model bias for the DNA. The asymmetric unit of the structure consists of the protein molecule bound to the blunt end of a DNA 6/10-mer, which is composed of a six-base strand (5'-GTCGTC-3') and a ten-base strand (3'-CAGCAGGGCA-5'), resulting in a six-base-pair duplex with a four-base single-stranded overhang. In the crystal structure, the bases of the overhang reciprocally pair to yield a doubly nicked pseudo-hexadecamer primarily B-form DNA molecule. The pairing between the single strands gives two standard (G-C) Watson-Crick pairs and two G(anti)-A(anti) mispairs. The mispairs reside in a G-C-rich environment and the three consecutive guanines on the 10-mer impart interesting structural features to the pseudo-hexadecamer, such as the preference for a guanine stack, stretching the C-G base pairs flanking the mispair to the point of loss of intra-base-pair hydrogen bonding. The DNA was designed for the purpose of comparison with a previous structure, which was determined in the same crystal lattice. In all of the authors' previous fragment-DNA complexes, the nucleotide at the blunt-ended 3'-hydroxyl was a purine. Consistent with the proposed mechanistic role of interactions with the 3'-hydroxyl in processive DNA synthesis by RT, it was found that a pyrimidine at this position in the DNA makes indentical interactions with the strictly conserved Gly191 and the main chain of Leu115 of MMLV RT.